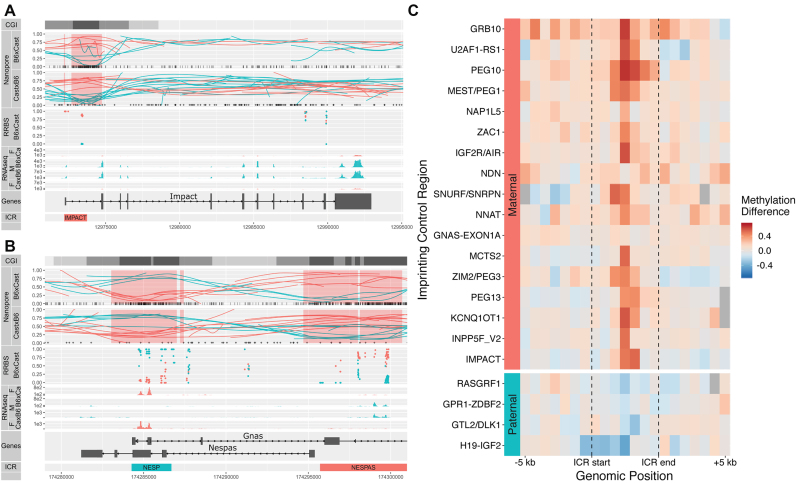
Figure 4.
Nanopore allele-specific methylation captures known differential methylation at ICRs. (A) Allelic methylation plot of maternally imprinted gene Impact displays a clear DMR at its ICR. Haplotyped RRBS data shows concordance with nanopore allelic methylation. Allele-specific RNA-seq coverage plots show monoallelic paternal expression. CGIs are displayed in black, with CpG shores in dark grey and CpG Shelves in light grey. Nanopore: Vertical bars at the base of the B6Cast track denote CpG sites used for methylation calling, while ‘+’ signs at the base of the CastB6 track denote SNPs used for haplotyping. Highlighted red regions indicate DMRs detected by DSS. The maternal allele is shown in red and the paternal allele in cyan for all plots. (B) Allelic methylation plot as in A for the reciprocally imprinted genes Nespas and Gnas. RNA-seq gives very low expression and is not shown. (C) Heatmap of differences (maternal − paternal) in allelic methylation in relative-width bins along known ICRs. Regions are sorted in order of average methylation difference, with regions in the same imprinting cluster placed adjacent to each other. Regions without haplotyped calls for both alleles are shown in grey.