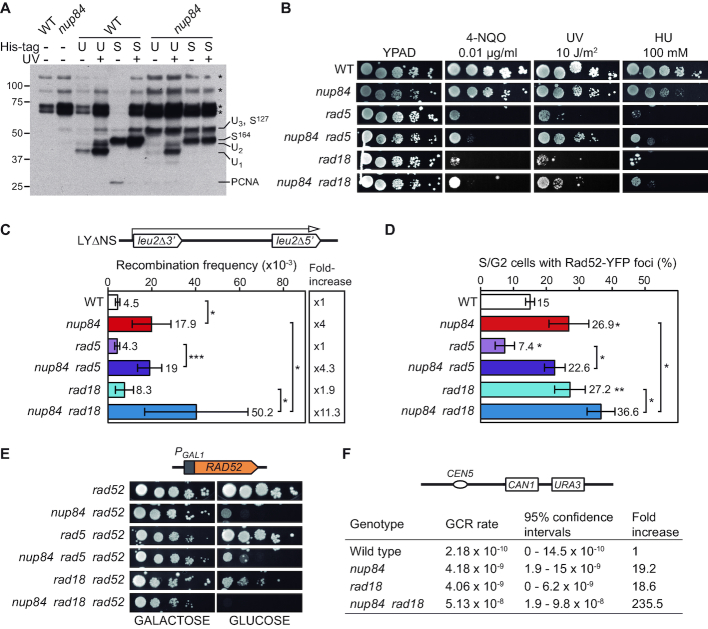
Figure 6.
Nup84 is required for appropriate post replication repair. (A) Pattern of PCNA modifications in wild-type (WT, BY4741) and nup84Δ (YDL116W) cells in untreated or UV-irradiated (150 J/m2) cultures. Ubiquitylated (U) and sumoylated (S) forms of endogenous PCNA were affinity purified from cells expressing His-tagged ubiquitin and Smt3 and detected by anti-PCNA Western blot. Migration of the different species as described in (35) and unspecific signals are indicated. U1, U2 and U3: mono- di- and tri-ubiquitylated PCNA species, respectively; S127 and S164: sumoylation on K127 and K164 residue, respectively; PCNA: unmodified PCNA; *: unspecific signal. (B) Sensitivity of wild type (WT, BY4741), nup84Δ (YDL116W), rad5Δ (YLR032W), nup84Δ rad5Δ (yHG132-13C), rad18Δ (yHG128-3D) and nup84Δ rad18Δ (yHG130-3D) to 4-NQO, UV and HU. 10-fold serial dilutions of exponentially growing cells are shown. (C) Recombination analysis of WT, nup84Δ, rad5Δ, nup84Δ rad5Δ, rad18Δ and nup84Δ rad18Δ cells described in A using the plasmid borne LYΔNS direct-repeat system (N ≥ 4). A scheme of the recombination system is shown. *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired Student's t-test) (D) Percentage of S/G2 cells with Rad52-YFP foci in WT, nup84Δ, rad5Δ, nup84Δ rad5Δ, rad18Δ and nup84Δ rad18Δ cells described in A (N ≥ 3). Statistical analyses as in B. (E) Growth in galactose and glucose-containing media of rad52Δ cells expressing wild type RAD52 under the control of the GAL1 promoter in combination with the nup84Δ, rad5Δ and rad18Δ mutations obtained by genetic crosses between strains BYR52α (transformed with pDML5) and yHG130-3D or yHG132-13C. (F) Gross chromosomal rearrangement (GCR) rates obtained by scoring Canr 5-FOAr cells in WT (yHG163-7A), nup84Δ (yHG163-5D), rad18Δ (yHG163-4C) and nup84Δ rad18Δ (yHG163-3A) cells (N ≥ 11). 95% confidence intervals for the median and fold change relative to WT are shown. A scheme of the system is depicted on top.