Figure 1.
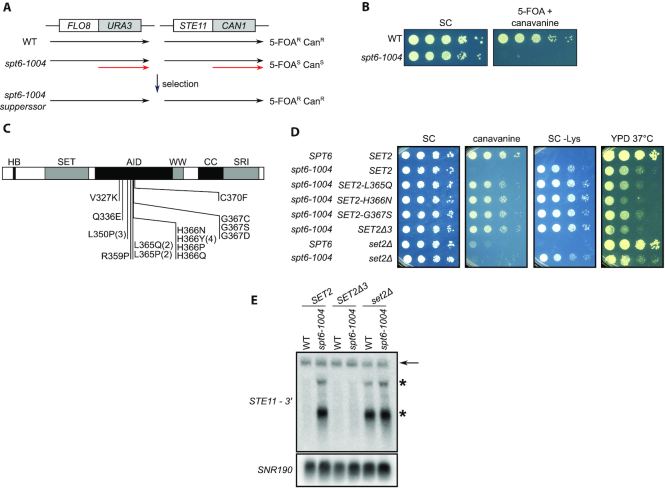
Isolation of mutations that suppress intragenic transcription in spt6-1004. (A) Shown at the top are diagrams of the two reporters and the selection used to isolate suppressor mutations. Shown below each are the transcripts made in wild-type (WT), spt6-1004 and spt6-1004 strains with a suppressor. In spt6-1004 mutants, the FLO8-URA3 reporter confers 5-FOA sensitivity and the STE11-CAN1 reporter confers canavanine sensitivity due to expression of the intragenic transcripts, shown in red. (B) Spot tests of cells grown at 34°C that show the selective conditions for the mutant selection. (C) A diagram of the Set2 protein that depicts the amino acid changes caused by the dominant SET2sup mutations. The numbers in parentheses indicate the number of times the same mutation was isolated if more than once. One isolate had two point mutations that coded for L365P and H366Y in the same protein. The letters above the rectangle indicate the identified domains in Set2: HB, histone binding domain; SET, the catalytic domain; AID, the autoinhibitory domain; the WW domain; CC, a coiled coil motif; SRI, the Set2 Rpb1 interacting domain. (D) Spot tests of cells grown at 34°C to assay suppression of intragenic transcription in spt6-1004 by SET2sup mutations using the STE11-CAN1 reporter. (E) Northern analysis of the STE11 gene, using a probe from the 3′ region of STE11, to assay the suppression of intragenic transcription in spt6-1004 by a SET2sup mutation. In wild-type, there is a single full-length STE11 transcript (denoted by the arrow), while in the spt6-1004 mutant, there are two intragenic transcripts (denoted by the asterisks) in addition to the full-length transcript. SNR190 served as the loading control.