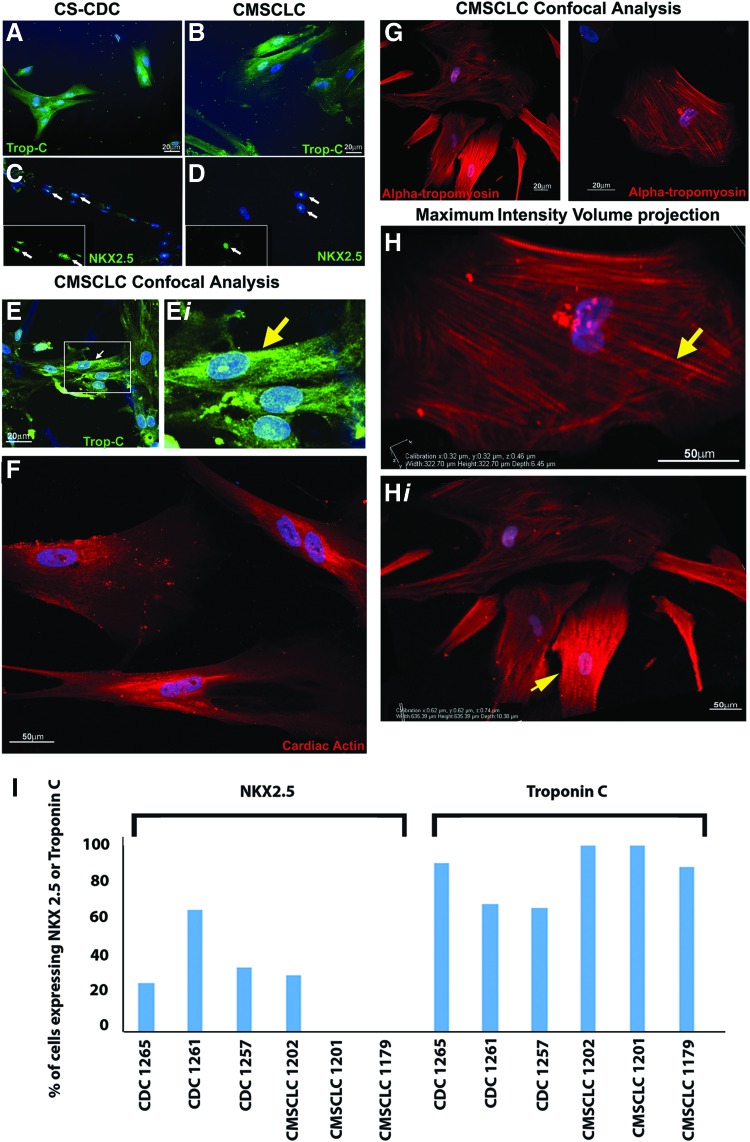
FIG. 6.
Characterization of cardiac differentiation potential of CDCs and CMSCLC. Representative images of CDC (n = 3 donors) and CMSCLC (n = 4) donors at 14 days under cardiac differentiation culture conditions show that some of both CDC (A) and (B) CMSCLC differentiated cells have a more mature cardiac morphology and express troponin C (green-Alexa Fluor 488). While other cells from the same patient samples cultured under the same conditions but stained for expression of NKX2.5 have a more primitive rounded morphology and express nuclear NKX2.5 (NKX2.5 green-Alexa Fluor 488) in differentiated CDC (C) and differentiated CMSCLC culture (D). White arrows in (C, D) indicate cells shown in inserts of NKX2.5 staining without the DAPI overlay to shown the nuclear localization of NKX2.5. (E–H) Representative images of CMSCLC (n = 4 donors) differentiated for 3 weeks and analyzed by confocal microscopy. (E) Maximum intensity projections of differentiated CMSCLC expressing troponin C (green-Alexa Fluor 488). (Ei) Higher magnification image demonstrates early striations, indicated by yellow arrow. (F) Maximum intensity volume projection of differentiated CMSCLC labeled with cardiac actin (red-Alexa Fluor 594). (G) Maximum intensity volume projection of differentiated CMSCLC labeled with alpha-tropomyosin (red-Alexa Fluor 594). (H, Hi) Maximum intensity volume projection demonstrates striated pattern of alpha-tropomyosin expression, indicated by yellow arrows. For all images, nuclei labeled with DAPI-blue. Scale bars were either 20 or 50 μm as indicated. (I) Bar chart showing percentage of cells expressing NXK2.5 or troponin C, n = 3 for both CDC and CMSCLC cultures analyzed. Color images are available online.