Significance
CCR2 is an important immune system protein that is implicated in many immune and inflammatory diseases, including cancer. Numerous clinical and preclinical trials are attempting to target CCR2 to find an effective therapy. To support drug discovery efforts, we use atomic simulations and Markov-state models to investigate the conformational landscape of CCR2, information that is unattainable by experimental methods.
Keywords: CC chemokine receptor 2, CCR2, GPCR, Markov-state model, molecular dynamics simulations
Abstract
CC chemokine receptor 2 (CCR2) is a part of the chemokine receptor family, an important class of therapeutic targets. These class A G-protein coupled receptors (GPCRs) are involved in mammalian signaling pathways and control cell migration toward endogenous CC chemokine ligands, named for the adjacent cysteine motif on their N terminus. Chemokine receptors and their associated ligands are involved in a wide range of diseases and thus have become important drug targets. CCR2, in particular, promotes the metastasis of cancer cells and is also implicated in autoimmunity-driven type-1 diabetes, diabetic nephropathy, multiple sclerosis, asthma, atherosclerosis, neuropathic pain, and rheumatoid arthritis. Although promising, CCR2 antagonists have been largely unsuccessful to date. Here, we investigate the effect of an orthosteric and an allosteric antagonist on CCR2 dynamics by coupling long-timescale molecular dynamics simulations with Markov-state model theory. We find that the antagonists shift CCR2 into several stable inactive conformations that are distinct from the crystal structure conformation and disrupt a continuous internal water and sodium ion pathway, preventing transitions to an active-like state. Several metastable conformations present a cryptic drug-binding pocket near the allosteric site that may be amenable to targeting with small molecules. Without antagonists, the apo dynamics reveal intermediate conformations along the activation pathway that provide insight into the basal dynamics of CCR2 and may also be useful for future drug design.
The signaling axis of CC chemokine receptor 2 (CCR2) and its endogenous ligand, CCL2, is a notable therapeutic target due to its association with numerous diseases, including cancer, autoimmunity-driven type-1 diabetes, diabetic nephropathy, multiple sclerosis, asthma, atherosclerosis, neuropathic pain, and rheumatoid arthritis (1–3). Despite much effort that has been devoted to clinical and preclinical trials, a successful antagonist has yet to be developed (4–7). Before a full-length crystal structure of CCR2, several studies used homology modeling and docking to gain insights into the structure and dynamics of the protein and its associated ligands or small molecule drugs (8–10). However, recently CCR2 was crystallized for the first time (11), opening up new opportunities for rational drug design.
As with most G-protein coupled receptors (GPCRs), chemokine receptors transmit signals across cell membranes by means of extracellular ligand and intracellular G-protein binding. Distinct conformational states of the receptor are necessary for chemokine/ligand binding, G-protein binding, activation, inactivation, and signal transmission (12–14). GPCRs are no longer considered to be simple on/off molecular switches—instead, they assume a wide range of conformational states, including ligand-specific states, intermediate states, and states that allow for basal (apo) signaling without ligands bound (13, 15–21). Ligands and small molecule drugs may shift the equilibrium of the receptor’s conformational states toward particular states. Effective small molecule antagonists that inhibit CCR2 signaling, potentially by shifting the receptor equilibrium toward inactive conformational states, are desired for treatment of diseases that involve the CCR2/CCL2 axis. Key challenges are to characterize the basal dynamics of CCR2 and to understand how current antagonistic small molecule drugs modulate these dynamics. While crystal structures provide valuable snapshots of proteins and protein complexes, they lack the ability to reveal dynamics at the atomic level. Starting with the newly resolved crystal structure of CCR2 [Protein Data Bank (PDB) ID: 5T1A], we performed multimicrosecond all-atom explicitly solvated molecular dynamics (MD) simulations of the receptor in a lipid bilayer in unbound (apo) and dual-antagonist–bound (holo) states (Fig. 1). The two antagonists were cocrystallized with CCR2: the orthosteric antagonist, BMS-681, and the allosteric antagonist, CCR2-RA-[R]. Each system was simulated in triplicate on Anton 2 (22) for a total of 260 s (SI Appendix, Table S1 and Fig. S1).
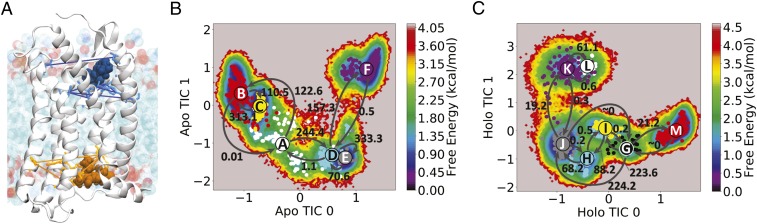
Fig. 1.
MD simulations of CCR2 in a lipid bilayer were performed in apo and holo CCR2. (A) Sets of residue pairs surrounding the two ligand-binding pockets were used with TICA (SI Appendix). The protein is shown in a white cartoon. Lipids are teal, red, and blue. The orthosteric and allosteric ligands are shown in blue and orange, respectively, with interresidue pair distances denoted by similarly colored lines. (B and C) The free-energy and maximum-likelihood HMMs of (B) apo and (C) holo CCR2, projected onto the first two TICA components. Coarse-grained states are labeled and colored. Transition rates between macrostates are represented by arrows reported in units of ms−1.
While long-timescale (tens of microseconds) simulations are useful for analyzing sequential conformational changes, simulations are generally unable to directly probe timescales of biological interest (milliseconds to seconds) (23). One way to bridge this timescale gap is to couple MD simulations with Markov-state model (MSM) theory (24–32) (described in SI Appendix, Methods). Integrating MD simulations with MSMs allowed us to extend the reach of simulated timescales and identify key differences in the conformational ensembles and dominant slow motions of apo and holo CCR2 (Fig. 1). We find that the antagonists disrupt a continuous internal water and sodium ion pathway, preventing transitions to an active-like state, and shift CCR2 into several stable states that are distinct from the crystal structure conformation, three of which present a cryptic druggable pocket. Without antagonists, intermediate conformations with active-state conformational signatures shed light on the apo dynamics of CCR2 and may also be useful for future drug design.
Results and Discussion
To compare the conformational landscapes of apo and holo CCR2 we ran all-atom MD simulations totaling 260 s on Anton 2 (22, 33). For one MD replicate of the holo system, we observed the orthosteric drug dissociate from the protein. Analyzing the conformations before and after ligand dissociation yields a first glimpse of the allosteric effect of the remaining antagonist on the protein dynamics and provides a starting point for future rounds of adaptive sampling to obtain robust dissociation statistics (not pursued here). To extend the analysis beyond a dissociation event and connect to longer-timescale phenomena, MSMs were constructed from the trajectories (Fig. 1 B and C). The variational approach for conformation dynamics (34), specifically time-structure–based independent component analysis (TICA) (35, 36), was used to perform dimensionality reduction for the MSMs and identify the features and collective variables [time-structure–based independent components (TICs)] that best represent the dominant slow motions. The MSMs create human-interpretable models that we use to interrogate the conformational and kinetic differences between the two ensembles to derive understandings about the mechanisms underlying the effects of CCR2 antagonism. Further methodological details are provided in Materials and Methods and SI Appendix.
Comparison of the CCR2 Conformational Ensemble with Other Class A GPCRs.
We compare representative states from the apo and holo conformational ensemble with other class A GPCRs to establish similarities within the class. We find that the states of apo CCR2 have conformational signatures found in the active or intermediately active states of GPCRs, suggesting that these states are on a pathway toward activation. Holo CCR2 diverges from the crystal structure to form distinct states that expose putative drug-binding pockets and reveal the effect of antagonists on receptor dynamics. The most populated holo macrostate, J, is not representative of the crystal structure as it deviates 10.8 Å from the crystal structure conformation (Fig. 1C).
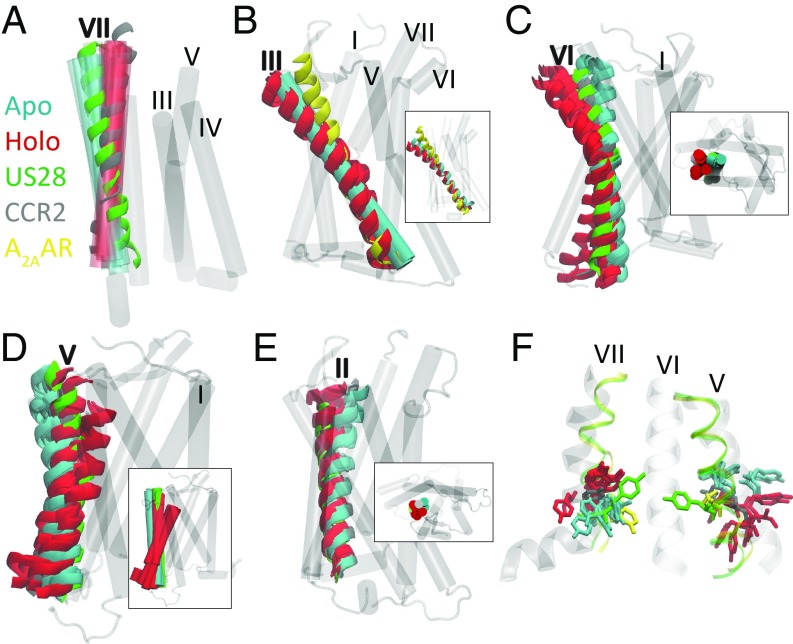
We evaluate the metastable states by comparing helical conformational signatures and conserved groups of structurally neighboring amino acids called “microswitches.” These include NPxxY (Tyr- 3057.53), DRY (Arg- 1383.50), Tyr- 2225.58, sets of residues in the orthesteric and allosteric binding sites, and the chemokine and G-protein binding pockets to the inactive crystal structure of CCR2 that we used in this study (PDB ID: 5T1A); an intermediately active crystal structure of a class A GPCR, A2AAR [PDB ID: 2YDO (37); 25% sequence identity to CCR2]; the active crystal structure of a class A GPCR, US28 [PDB ID: 4XT3 (38); 30% sequence identity to CCR2]; and three other chemokine receptors, CCR5 [PDB ID: 4MBS (39)], CCR9 [PDB ID: 5LWE (40)], and CXCR4 [PDB ID: 4RWS (41)]. Signatures of an active GPCR state include (i) the inward shift of the intracellular part of helix VII toward the helical bundle, (ii) the outward shift of the intracellular part of helix VI in concert with helix V, (iii) the upward shift and lateral movement of helix III, and (iv) the rearrangements of conserved microswitches (15). According to these metrics, the starting crystal structure of CCR2 is in an inactive conformation (11), the crystal structure of A2AAR is in an intermediately active conformation, and the crystal structure of US28 is in an active conformation.
Apo macrostates show an active-like inward shift of the intracellular part of helix VII toward the helical bundle.
All of the apo macrostates exhibit an active-state hallmark (Fig. 2A): The intracellular end of helix VII tilts slightly inward toward the center of the helical bundle. More prominently, the extracellular end of helix VII tilts outward, resembling the active conformation of US28. The holo macrostates show the opposite: The intracellular end of helix VII tilts slightly outward and the extracellular end of helix VII tilts inward, remaining in the crystal structure conformation.
Fig. 2.
Apo and holo macrostates are compared with the active crystal structure of US28 (green, PDB ID 4XT3) or the active crystal structure of A2AAR (yellow, PDB ID 2YDO) and the inactive crystal structure of CCR2 (gray, PDB ID 5T1A). (A) Helix VII of apo macrostates resembles the active conformation of US28; holo macrostates resemble the CCR2 crystal structure. (B) The conformation of helix III in apo macrostates subtly resembles active A2AAR; holo macrostates are tilted away from the center of the helical bundle. (C and D) Helices V and VI of apo macrostates straighten or tilt in toward the center of the protein, similar to the active conformation of US28 and the inactive CCR2 crystal structure; helices V and VI of several holo macrostates tilt away from the binding sites, accessing more active-like conformations than the apo macrostates or even the active state of US28. (E) Helix II in the apo macrostates shifts inward; in the holo macrostates it shifts outward. (F) In licorice are conserved motifs TYR- 3057.53 and TYR- 2225.58. All six apo metastable states assume a new conformation for TYR- 3057.53, pointing intracellulary and in a similar conformation to active A2AAR. Six of the seven holo metastable states have TYR- 3057.53 in the same conformation as the equilibrated crystal structure. Postligand-dissociation holo state L assumes a new position of TYR- 3057.53, more similar to the dominant apo conformation. Apo metastable states sample a narrower range of conformations for TYR- 2225.58 than holo.
Holo macrostates, not apo, show an active-like outward shift of the intracellular part of helix VI in concert with helix V.
Helices V and VI in the apo macrostates are not in an active conformation. Instead, it is the holo macrostates that have the intracellular end of helices V and VI tilting outward to resemble the active conformation (Fig. 2 C and D), suggesting that neither apo nor holo macrostates are in an exclusively inactive or active conformational state, despite starting from a particularly inactive crystal structure. Due to this outward motion of helix VI, holo macrostates K and L exhibit a more open G-protein binding site compared with holo macrostate G which is more closed (Fig. 1 B and C). The root mean square fluctuation of the allosteric ligand is larger in macrostates K and L, indicating that the inactive (inward) conformation of helix VI may play a critical role in stabilization of the allosteric ligand (SI Appendix, Fig. S2).
Apo macrostates show an active-like upward shift and lateral movement of helix III.
Apo macrostates also resemble the active conformation by the slight upward shift of helix III, unlike holo macrostates, which remain in a position similar to the inactive crystal structure (Fig. 2B).
The rearrangements of conserved microswitches suggest that apo macrostates resemble active states, and holo macrostates resemble inactive states.
NPxxY motif (Tyr- 3057.53).
In the inactive conformation of GPCRs, Tyr- 3057.53 points toward helix I, II, or VIII (in CCR2, it points toward II), and in the active state Tyr- 3057.53 points toward the middle axis of the helical bundle (15). Each apo macrostate shows Tyr- 3057.53 pointing downward into the intracellular (G-protein) binding pocket (Fig. 2F). This positioning of Tyr- 3057.53 matches the intermediately active conformation of A2AAR, which also points down. It does not match the active conformation in US28 that points up and is also distinct from the inactive crystal structure of CCR2. In six of the seven holo macrostates, Tyr- 3057.53 is stabilized in the inactive state and matches the inactive CCR2 crystal structure conformation as well as the inactive chemokine receptor crystal structures of CCR5 and CCR9.
The holo macrostate in which Tyr- 3057.53 is not stabilized in the inactive conformation is accessed after the orthosteric ligand dissociates (state L, Figs. 1C and 2F); the allosteric pocket residues rearrange and Tyr- 3057.53 assumes a downward conformation similar to the apo states and CXCR4. These concerted events may indicate allosteric cross-talk between the chemokine binding site and the G-protein binding site.
The microswitch residue Trp- 2566.48 and the interaction of the DRY motif (Arg- 1383.50) with Tyr- 2225.58.
Apo and holo macrostates both maintain the same chi angle of the conserved microswitch residue Trp- 2566.48 which describes an active GPCR when it switches from gauche to trans conformation and facilitates the interaction of Tyr- 2225.58 and Tyr- 3057.53. In the CCR2 crystal structure and the crystal structures of chemokine receptors CCR5 and CXCR4, Trp- 2566.48 points upward and extends into the helical core. Each apo macrostate shows Trp- 2566.48 in a single conformation pointing toward helix III (SI Appendix, Fig. S3). In the holo macrostates, Trp- 2566.48 accesses three distinct conformations: one resembling the crystal structure but with the helix shifted slightly outward from the helical core, another one that laterally twists toward helix V, and another conformation that points down into the helical core toward the G-protein binding site (SI Appendix, Fig. S3). This third conformation, in which the orthosteric ligand has dissociated, represented by the holo macrostate L, further suggests cross-talk between the chemokine binding site and the rest of the protein.
The interaction of these two tyrosines and Arg- 1383.50 also characterizes an active-state GPCR (42). In the inactive crystal structure of CCR2, Tyr- 2225.58 points toward lipids, sterically blocked by Phe- 2466.38 from interaction with Arg- 1383.50 and Tyr- 3057.53 (11). In apo macrostates, Tyr- 2225.58 remains pointed toward the lipids, never swiveling around to interact with Arg- 1383.50 or Tyr- 3057.53 as occurs in activated GPCR states (Fig. 2F). Holo macrostates actually show increased range of motion of Tyr- 2225.58, diverging from the crystal structure and stabilizing in unique intermediate conformations. The steric obstruction from Phe- 2466.38 is alleviated in both apo and holo macrostates, as Phe- 2466.38 swings outward and points toward the lipids. The conformations of these microswitch residues indicate that both apo and holo macrostates are sampling different conformations.
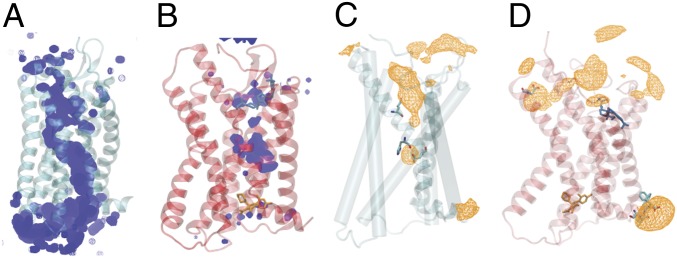
Formation of continuous water pathway suggests movement of apo toward activation.
Internal water molecules, which may influence conformational changes in GPCRs by interfering with hydrogen-bonding networks of the receptor’s backbone and side chains, are postulated to be an integral part of receptor activation in GPCRs (43–46). Work in other GPCRs has additionally shown that activation can allow water and sodium ions to flow through the GPCR core (47). Furthermore, it has been shown that the activation of GPCRs is voltage sensitive (48). Our simulations enable the direct visualization of water and sodium ion density in both CCR2 ensembles.
A continuous internal water pathway forms in apo CCR2 (Fig. 3A). The antagonists disrupt this water pathway, slowing the rate of water entry into and egress out of the protein core (Fig. 3A). An analysis of the water occupancy per residue (SI Appendix, Fig. S4A) indicates that several of the high water occupancy residues (e.g., Asp- 361.26, Ser- 501.40, Glu- 2356.27, Lys- 2366.28, Glu- 3108.48, and Lys- 3118.49) may be exposed to more water in the apo simulations than in the holo simulations simply because the ligands have been removed and the water has access to the binding pockets. The other residues (e.g., Asp- 782.40, Tyr- 802.42, Asp- 882.50, Leu- 922.54, Ile- 932.55, Gly- 1273.39, Ile- 1283.40, Glu- 2917.39, and Phe- 3128.50) reside in the protein core, along the continuous pathway (Fig. 3A).
Fig. 3.
Ligands disrupt a continuous internal water and sodium ion pathway. (A and B) Average water density over a 50-s simulation of (A) apo (teal) and (B) holo (red). The orthosteric ligand is shown in blue and the allosteric ligand is shown in orange. (C and D) Total average sodium ion density in (C) apo and (D) holo. Highest-occupancy residues are depicted in cyan licorice and plotted in SI Appendix, Fig. S4.
Class A GPCRs possess a conserved sodium binding site at Asp2.50 corresponding to Asp- 88 in CCR2 (49). The role of sodium is thought to contribute to the mechanism of receptor activation (50–52). In particular, dynamics of activation were previously hypothesized to impinge upon the sodium binding pocket, eventually leading to ion permeation from the sodium binding site into the cytosol (51). A sodium ion occupancy per residue analysis (Fig. 3C and SI Appendix, Fig. S4B) indicates that, while no sodium permeation events into the cytosol were observed in the apo trajectories, ions interact with sodium binding-site residues Asp- 882.50, Glu- 2917.39, and His- 2977.45. In holo CCR2, sodium does not interact with binding-site residues, preventing the possibility of a permeation event (Fig. 3D and SI Appendix, Fig. S4B).
Effects of Antagonist Binding on CCR2 Dynamics.
Comparisons of the apo and holo MSMs elucidate the effects of antagonists on CCR2 dynamics. Notably, apo relaxation timescales are an order of magnitude less than holo relaxation timescales (SI Appendix, Table S2), indicating that the antagonists greatly perturb CCR2 dynamics. The motions described by apo and holo TIC 0 represent the most striking difference between the two systems’ dynamics. In the apo MSM, the interresidue distances most closely correlated with apo TIC 0 are all a part of the allosteric (G-protein) binding pocket, whereas in the holo MSM, the interresidue distances most closely correlated with holo TIC 0 are all a part of the orthosteric (chemokine) binding pocket (SI Appendix, Fig. S5).
Apo TIC 1 represents the flipping of Trp- 982.60 into the orthosteric drug binding site (SI Appendix, Figs. S5C and S7 A and B). In the crystal structure Trp- 982.60 packs with the trisubstituted cyclohexane of the orthosteric antagonist, BMS-681 (11). Without the presence of this ligand, Trp- 982.60 assumes three distinct positions. The Trp- 982.60 conformation in the cluster at the neutral TIC (boxed in yellow; SI Appendix, Fig. S7 A and B) most closely resembles the conformation of Trp- 982.60 in the active GPCR US28, which is shifted slightly up and in toward the helical core in comparison with the CCR2 crystal structure. The two other conformations are found at the extreme ends of apo TIC 1 in densely populated free-energy wells. Of these two conformations, state F assumes the most dramatic conformation and protrudes into the chemokine binding site (SI Appendix, Fig. S6). In the holo macrostates there is markedly less intrusion into the binding pocket due to the presence of the ligand.
Holo TIC 1 represents the concerted movement of five pairs of residues in the orthosteric ligand binding site during orthosteric ligand dissociation (SI Appendix, Figs. S5D and S7 C and D). The separation projected in the first two TICs (SI Appendix, Fig. S7D) is divided into clusters of frames that occur before (white clusters), during (gray), and after (black) dissociation. The residue pairs identified by TICA that contribute to holo TIC 1 and this ligand dissociation (SI Appendix, Fig. S5D) were confirmed by analyzing the original simulation data. The key changes are the change in distance between Tyr- 491.39–Thr- 2927.40, Trp- 982.60–Tyr- 1203.32, and Ser- 501.40–Tyr- 2596.51 and in the chi angle of Glu- 2917.39. Notably, four of these residues (Tyr- 491.39, Trp- 982.60, Tyr- 1203.32, and Thr- 2927.40) not only are involved in binding to the cocrystallized orthosteric antagonist BMS-681 and/or CCL2 binding, but also are critical for GPCR activation (53, 54).
The positioning of the orthosteric ligand and the conformation of Trp- 982.60 are closely linked (SI Appendix, Figs. S7C and S8). After ligand dissociation, in holo states K and L (purple and white, respectively; SI Appendix, Figs. S7C and S8), Trp- 982.60 turns toward helix III, bending slightly inward toward the chemokine binding site. Before ligand dissociation, Trp- 982.60 has two distinct conformations. In the first conformation (states I and G, yellow and black; SI Appendix, Figs. S7C and S8), the ligand positions itself between helices I and VII, in the same conformation as the crystal structure. Trp- 982.60 is constrained in a downward position, pointing intracellularly, also resembling the CCR2 crystal structure conformation and the crystal structure of chemokine receptor CCR5. In the second conformation (states H and J, cyan and gray; SI Appendix, Figs. S7C and S8), Trp- 982.60 flips up and out of the binding pocket, pointing extracellularly, and the ligand moves between helices I and II. This conformation of Trp- 982.60 more closely resembles CCR9 (SI Appendix, Fig. S9). The third conformation of Trp- 982.60 is found in state M (red; SI Appendix, Figs. S7C and S8) and is the most prominent position of the residue as it extends deeper into the chemokine binding site toward helix III. In this case, the ligand interacts with helices II, IV, and V, and there are no transitions from this state to a dissociated state.
As in the apo MSM, the absence of the orthosteric ligand causes a shift in the position of Trp- 982.60. In the holo simulations shown in SI Appendix, Fig. S10, the dissociation event is preceded by a doubling of the distance between Trp- 982.60 and Tyr- 1203.32, and 3 s after dissociation the distance returns to its previous 0.4 nm. This increase in distance may be required for the ligand to begin the process of dissociating. Another drastic change during the dissociation event is the switch of Glu- 2917.39 from a constrained chi angle of −50° to −100° to an unconstrained chi angle (SI Appendix, Fig. S11). After dissociation, this angle more closely resembles the conformation in all apo simulations. Glu- 2917.39 is a key mediator of many CCR2 antagonists (55), but there is no direct interaction between Glu- 2917.39 and the orthosteric antagonist in the CCR2 crystal structure (41). That the conformation of Glu- 2917.39 switches after dissociation suggests that Glu- 2917.39 is involved in ligand stabilization despite not directly interacting with the ligand.
In the CCR2 crystal structure, there is a hydrogen bond between Tyr- 491.39 and Thr- 2927.40. The gamma-lactam secondary exocyclic amine of the orthosteric ligand forms a hydrogen bond with the hydroxyl of Thr- 2927.40, and the carbonyl oxygen of the gamma-lactam forms a hydrogen bond with Tyr- 491.39. During simulation, the distance between Tyr- 491.39 and Thr- 2927.40 remains stable until 3 s after the ligand dissociates, when it begins fluctuating (SI Appendix, Fig. S12). This suggests that the orthosteric ligand dissociation breaks the hydrogen bond between these key ligand binding residues. This motion is captured in the holo MSM: The separation of the two residues is exemplified between states H (predissociation) and L (postdissociation) in SI Appendix, Fig. S12.
Finally, the faster dominant motions (TICs 2, 3, and 4) in the holo MSM consist of rearrangements in the allosteric ligand binding site, suggesting that an allosteric rearrangement must first happen for the orthosteric ligand to dissociate. Further evidence for this is the observed correlated motion of the downward flip of the conserved residue Tyr- 3057.53 in the G-protein binding site with the dissociation of the orthosteric ligand from the chemokine binding site.
Overall, holo macrostates show more helical tilting and binding-site expansion, which increases the solvent-accessible surface area (SASA) compared to that in the crystal structure and the apo macrostates. However, the apo simulations overall have greater residue fluctuation, suggesting that the antagonist ligands dampen CCR2 dynamics (SI Appendix, Fig. S13).
Opening of a Cryptic Druggable Pocket.
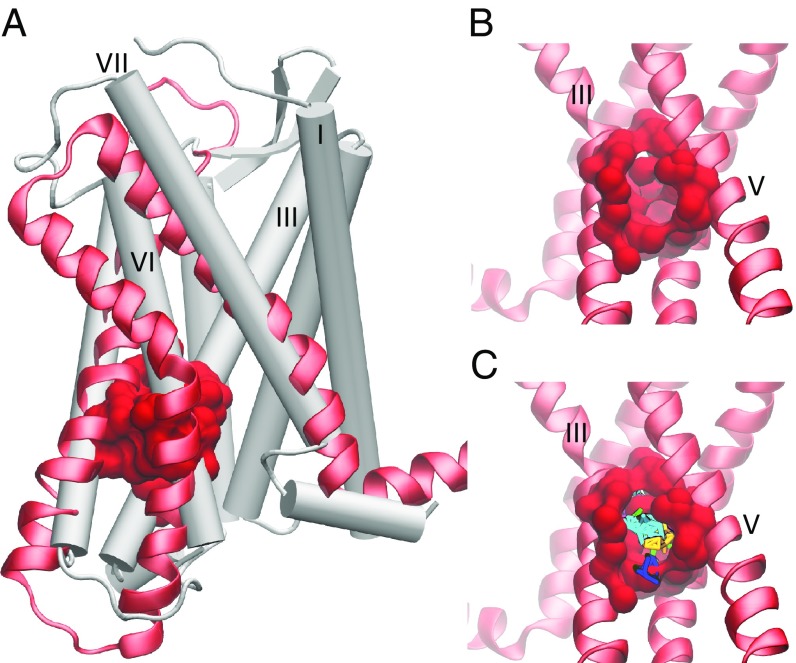
A dramatic expansion of the extracellular (chemokine) binding site is exhibited in the holo macrostates. The expansion is caused by a pronounced outward tilting of helix VI and slight outward tilting of helix II in the holo macrostates, whereas the apo macrostates show the opposite, with a slight inward tilting of both helices VI and II (Fig. 2 C and E). The intracellular (G-protein) binding site also enlarges in the holo macrostates due to the outward shift of the intracellular ends of helices V and VI, but remains obstructed in all apo and holo states. In the crystal structure, this obstruction occurs by the interaction of Arg- 1383.50 with Asp- 1373.49 and with Thr- 772.39 (11), which are maintained throughout all of the simulations. The outward movement of the intracellular end of helix VI and the movement of helix V toward helix VI in states L, J, and H in the holo MSM create a putative site for allosteric antagonists; this pocket also transiently appears in the apo simulations (Fig. 4). Computational solvent mapping (56) of this site indicates that the pocket presents surfaces that are amenable to ligand binding due to its ability to bind clusters of multiple different drug-like probes (Fig. 4C). The pocket can be accessed through the lipid bilayer between helices IV and V, or from the G-protein binding site, as a deeper extension of the current allosteric binding site of CCR2-RA-[R], and may be useful for rational drug design or modification of current antagonists.
Fig. 4.
A putative allosteric drug binding pocket is revealed by three holo macrostates. (A) A comparison of the CCR2 crystal structure (white cartoon) with helices V, VI, and VII (red new cartoon) of one holo macrostate. The pocket is shown in red surface. (B) A closer view of the pocket from the other side of the protein, between helices III and V. (C) Small organic probes used for computational fragment mapping are multicolored.
Conclusions
To characterize the basal dynamics of CCR2 and understand how small molecule antagonists modulate these dynamics, we coupled long-timescale atomic simulations and MSM theory to compare the metastable states accessed by apo and holo CCR2 in its native membrane-embedded form.
Antagonists perturb CCR2 dynamics and kinetics and are associated with distinct residue rearrangement and key motions. Several intermediate states reveal a cryptic binding site that could be targeted with small molecule inhibitors. In a previous study (57), cryptic pockets predicted with MSM theory were experimentally confirmed and the results suggest that this methodology can successfully be used to guide drug discovery efforts.
Without antagonists, CCR2 is able to access other distinct metastable states that are likely sampling along an activation pathway. These intermediate states inform on the basal dynamics of CCR2 and may be useful for modification of previously unsuccessful drugs.
Materials and Methods
See SI Appendix, Methods for full description of materials and methods. MD trajectories and MSM construction scripts are available for download (33).
System Preparation and Molecular Dynamics Simulations.
Two systems were simulated for a total of 260 s: CCR2 holo, with both cocrystallized antagonist ligands bound, and CCR2 apo, without ligands bound. CCR2-RA-[R] and BMS 681 (11) were removed to build the apo system. Each all-atom system is embedded in a 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) bilayer, explicitly solvated with transferrable intermolecular potential with 3 points (TIP3P) and simulated with 150 mM NaCl, at pH 7.4, at 310 K and 1 bar. The initial coordinates were taken from the experimental crystal structure (11).
Building the MSMs.
The MSMs were built with PyEMMA version 2.5.4 (58), selected based on implied timescale plots (SI Appendix, Fig. S14) and Chapman–Kolmogorov tests (SI Appendix, Figs. S15 and S16), and coarse grained with hidden Markov models (HMMs). Representative structures were selected from each macrostate by taking the centroid of the most populated microstate (SI Appendix, Figs. S17–S19).
Supplementary Material
Acknowledgments
We thank Tracy Handel and Irina Kufareva for their valuable input and initial modification of CCR2 coordinates. We thank Frank Noe and Cecilia Clementi for their useful discussions regarding MSM construction. We also thank the organizers and participants of the 2018 Workshop on Free Energy Methods, Kinetics and Markov State Models in Drug Design for helpful input. This work was supported in part by the Director’s New Innovator Award Program NIH Grant DP2-OD007237, the National Biomedical Computation Resource NIH Grant P41-GM103426, and the National Science Foundation through The Extreme Science and Engineering Discovery Environment (XSEDE) supercomputing resources provided via Award TG-CHE060073 (to R.E.A.). C.T.L. also acknowledges support from the NIH Molecular Biophysics Training Program (Grant T32-GM008326). Anton 2 computer time was provided by the Pittsburgh Supercomputing Center (PSC) through Grant R01GM116961 from the NIH. The Anton 2 machine at PSC was generously made available by D. E. Shaw Research.
Footnotes
Conflict of interest statement: R.E.A. has equity interest in, and is a cofounder and on the scientific advisory board of Actavalon, Inc.
This article is a PNAS Direct Submission.
Data deposition: MD trajectories along with MSM construction and other analysis scripts have been deposited at the UC San Diego Library Digital Collections (https://doi.org/10.6075/J0QZ289Q).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814131116/-/DCSupplemental.
References
- 1.Ben-Baruch A. The multifaceted roles of chemokines in malignancy. Cancer Metastasis Rev. 2006;25:357–371. doi: 10.1007/s10555-006-9003-5. [DOI] [PubMed] [Google Scholar]
- 2.O’Connor T, Borsig L, Heikenwalder M. CCL2-CCR2 signaling in disease pathogenesis. Endocr Metab Immune Disord Drug Targets. 2015;15:105–118. doi: 10.2174/1871530315666150316120920. [DOI] [PubMed] [Google Scholar]
- 3.Solomon M, Balasa B, Sarvetnick N. CCR2 and CCR5 chemokine receptors differentially influence the development of autoimmune diabetes in the NOD mouse. Autoimmunity. 2010;43:156–163. doi: 10.3109/08916930903246464. [DOI] [PubMed] [Google Scholar]
- 4.Scholten DJ, et al. Pharmacological modulation of chemokine receptor function. Br J Pharmacol. 2012;165:1617–1643. doi: 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7:28697–28710. doi: 10.18632/oncotarget.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solari R, Pease JE, Begg M. Chemokine receptors as therapeutic targets: Why aren’t there more drugs? Eur J Pharmacol. 2015;746:363–367. doi: 10.1016/j.ejphar.2014.06.060. [DOI] [PubMed] [Google Scholar]
- 7.Horuk R. Chemokine receptor antagonists: Overcoming developmental hurdles. Nat Rev Drug Discov. 2009;8:23–33. doi: 10.1038/nrd2734. [DOI] [PubMed] [Google Scholar]
- 8.Shahlaei M, Fassihi A, Papaleo E, Pourfarzam M. Molecular dynamics simulation of chemokine receptors in lipid bilayer: A case study on C-C chemokine receptor type 2. Chem Biol Drug Des. 2013;82:534–545. doi: 10.1111/cbdd.12179. [DOI] [PubMed] [Google Scholar]
- 9.Chavan S, Pawar S, Singh R, Elizabeth Sobhia M. Binding site characterization of G protein-coupled receptor by alanine-scanning mutagenesis using molecular dynamics and binding free energy approach: Application to C-C chemokine receptor-2 (CCR2) Mol Divers. 2012;16:401–413. doi: 10.1007/s11030-012-9368-z. [DOI] [PubMed] [Google Scholar]
- 10.Kothandan G, Gadhe CG, Cho SJ. Structural insights from binding poses of CCR2 and CCR5 with clinically important antagonists: A combined in silico study. PLoS One. 2012;7:e32864. doi: 10.1371/journal.pone.0032864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR dynamics: Structures in motion. Chem Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 13.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q, Zhou M, Zhao L, Jiang H, Yang H. Dynamic states of the ligand-free class A G protein-coupled receptor extracellular side. Biochemistry. 2018;57:4767–4775. doi: 10.1021/acs.biochem.8b00146. [DOI] [PubMed] [Google Scholar]
- 15.Katritch V, Cherezov V, Stevens RC. Structure-function of the G-protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manglik A, et al. Structural insights into the dynamic process of beta(2)-adrenergic receptor signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik RU, et al. Detection of G protein-selective G protein-coupled receptor (GPCR) conformations in live cells. J Biol Chem. 2013;288:17167–17178. doi: 10.1074/jbc.M113.464065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci USA. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nygaard R, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bockenhauer S, Furstenberg A, Yao XJ, Kobil1 BK, Moerner WE. Conformational dynamics of single G protein-coupled receptors in solution. J Phys Chem B. 2011;115:13328–13338. doi: 10.1021/jp204843r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw DE, et al. International Conference for High Performance Computing, Networking, Storage and Analysis. IEEE; Piscataway, NJ: 2014. Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer; pp. 41–53. [Google Scholar]
- 23.Bowman GE, Voelz VA, Pande VS. Taming the complexity of protein folding. Curr Opin Struct Biol. 2011;21:4–11. doi: 10.1016/j.sbi.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swope WC, Pitera JW, Suits F. Describing protein folding kinetics by molecular dynamics simulations. 1. Theory. J Phys Chem B. 2004;108:6571–6581. [Google Scholar]
- 25.Singhal N, Snow CD, Pande VS. Using path sampling to build better Markovian state models: Predicting the folding rate and mechanism of a tryptophan zipper beta hairpin. J Chem Phys. 2004;121:415–425. doi: 10.1063/1.1738647. [DOI] [PubMed] [Google Scholar]
- 26.Malmstrom RD, Lee CT, Van Wart AT, Amaro RE. Application of molecular-dynamics based Markov state models to functional proteins. J Chem Theory Comput. 2014;10:2648–2657. doi: 10.1021/ct5002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaro RE, Mulholland AJ. Multiscale methods in drug design bridge chemical and biological complexity in the search for cures. Nat Rev Chem. 2018;2:0148. doi: 10.1038/s41570-018-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amaro RE, et al. Ensemble docking in drug discovery. Biophys J. 2018;114:2271–2278. doi: 10.1016/j.bpj.2018.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowman GR, Pande VS, Noe F. An Introduction to Markov State Models and Their Application to Long Timescale Molecular Simulation. Vol 797 Springer; The Netherlands: 2014. [Google Scholar]
- 30.Noé F, Horenko I, Schütte C, Smith JC. Hierarchical analysis of conformational dynamics in biomolecules: Transition networks of metastable states. J Chem Phys. 2007;126:155102. doi: 10.1063/1.2714539. [DOI] [PubMed] [Google Scholar]
- 31.Prinz JH, et al. Markov models of molecular kinetics: Generation and validation. J Chem Phys. 2011;134:174105. doi: 10.1063/1.3565032. [DOI] [PubMed] [Google Scholar]
- 32.SchÃŒtte Ch, Fischer A, Huisinga W, Deuflhard P. A direct approach to conformational dynamics based on hybrid Monte Carlo. J Comput Phys. 1999;151:146–168. [Google Scholar]
- 33.Taylor BC, Lee CT, Amaro RE. Data from “Structural basis for ligand modulation of the CCR2 conformational landscape.” UC San Diego Library Digital Collections. 2019 doi: 10.6075/J0QZ289Q. Available at . . Deposited September 17, 2018. [DOI] [PMC free article] [PubMed]
- 34.Noé F, Nüske F. A variational approach to modeling slow processes in stochastic dynamical systems. Multiscale Model Simul. 2013;11:635–655. [Google Scholar]
- 35.Schwantes CR, Pande VS. Improvements in Markov state model construction reveal many non-native interactions in the folding of NTL9. J Chem Theory Comput. 2013;9:2000–2009. doi: 10.1021/ct300878a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Hernandez G, Paul F, Giorgino T, De Fabritiis G, Noe F. Identification of slow molecular order parameters for Markov model construction. J Chem Phys. 2013;139:015102. doi: 10.1063/1.4811489. [DOI] [PubMed] [Google Scholar]
- 37.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burg JS, et al. Structural basis for chemokine recognition and activation of a viral G protein–coupled receptor. Science. 2015;347:1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Q, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor Maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oswald C, et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016;540:462–465. doi: 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- 41.Qin L, et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caliman AD, Swift SE, Wang Y, Miao Y, McCammon JA. Investigation of the conformational dynamics of the apo A2A adenosine receptor. Protein Sci. 2015;24:1004–1012. doi: 10.1002/pro.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jastrzebska B, Palczewski K, Golczak M. Role of bulk water in hydrolysis of the rhodopsin chromophore. J Biol Chem. 2011;286:18930–18937. doi: 10.1074/jbc.M111.234583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci USA. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choe H-W, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 46.Huang W, et al. Structural insights into -opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan S, Filipek S, Palczewski K, Vogel H. Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nat Commun. 2014;5:4733. doi: 10.1038/ncomms5733. [DOI] [PubMed] [Google Scholar]
- 48.Rinne A, Birk A, Bünemann M. Voltage regulates adrenergic receptor function. Proc Natl Acad Sci USA. 2013;110:1536–1541. doi: 10.1073/pnas.1212656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katritch V, et al. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci. 2014;39:233–244. doi: 10.1016/j.tibs.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan S, Vogel H, Filipek S. The role of water and sodium ions in the activation of the -opioid receptor. Angew Chem Int Ed Engl. 2013;52:10112–10115. doi: 10.1002/anie.201302244. [DOI] [PubMed] [Google Scholar]
- 51.Vickery ON, et al. Intracellular transfer of Na+ in an active-state G-protein-coupled receptor. Structure. 2018;26:171–180.e2. doi: 10.1016/j.str.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miao Y, Caliman AD, McCammon JA. Allosteric effects of sodium ion binding on activation of the M3 muscarinic G-protein-coupled receptor. Biophys J. 2015;108:1796–1806. doi: 10.1016/j.bpj.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berkhout TA, et al. CCR2: Characterization of the antagonist binding site from a combined receptor modeling/mutagenesis approach. J Med Chem. 2003;46:4070–4086. doi: 10.1021/jm030862l. [DOI] [PubMed] [Google Scholar]
- 54.Hall SE, et al. Elucidation of binding sites of dual antagonists in the human chemokine receptors CCR2 and CCR5. Mol Pharmacol. 2009;75:1325–1336. doi: 10.1124/mol.108.053470. [DOI] [PubMed] [Google Scholar]
- 55.Cherney RJ, et al. Synthesis and evaluation of cis-3,4-disubstituted piperidines as potent CC chemokine receptor 2 (CCR2) antagonists. Bioorg Med Chem Lett. 2008;18:5063–5065. doi: 10.1016/j.bmcl.2008.07.123. [DOI] [PubMed] [Google Scholar]
- 56.Kozakov D, et al. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowman GR, Bolin ER, Hart KM, Maguire BC, Marqusee S. Discovery of multiple hidden allosteric sites by combining Markov state models and experiments. Proc Natl Acad Sci USA. 2015;112:2734–2739. doi: 10.1073/pnas.1417811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scherer MK, et al. PyEMMA 2: A software package for estimation, validation, and analysis of Markov models. J Chem Theory Comput. 2015;11:5525–5542. doi: 10.1021/acs.jctc.5b00743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.