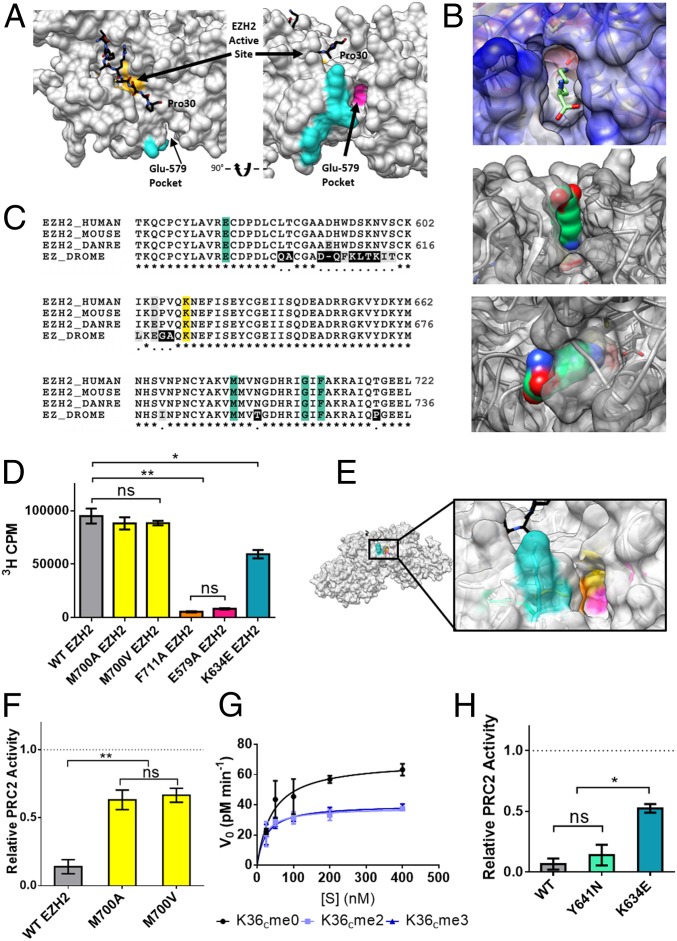
Fig. 4.
Role of the Glu-579 pocket on EZH2. (A) View of PRC2 with substrate H3 tail (black) bound in the active site (yellow). The cross-linked tryptic peptide is highlighted in cyan and the adjacent Glu-579 pocket highlighted in magenta (Protein Data Bank ID code 5HYN). (B, Top) an unmodified lysine side chain depicted as sticks modeled into the Glu-579 pocket in EZH2. (B, Middle and Bottom) a lysine side chain in space-filling view is shown modeled into the same pocket. (C) Sequence alignment of EZH2 shows conservation of pocket residues (teal) from fruit fly to human. DANRE, Danio rerio; DROME, Drosophila melanogaster. Cross-links were observed to the K634 residue of EZH2 (yellow), a residue notably mutated in some cases of Weaver syndrome. (D) WT or mutant PRC2 HMT activity as measured by scintillation counting on 200 nM unmodified mononucleosomes. Error bars represent SEM (n = 3). **P ≤ 0.01; *P ≤ 0.05; ns, not significant. (E) Zoom-in view of the Glu-579 pocket in EZH2 (Protein Data Bank ID code 5HYN), with pocket residues color-coded as follows: K634 in teal, M700 in yellow, F711 in orange, and E579 in pink. (F) WT or mutant PRC2 HMT activity as measured by scintillation counting on H3K36cme3 mononucleosomes, normalized to each enzyme’s activity on H3K36cme0 mononucleosome substrates (dotted line). Error bars represent SEM (n = 3). **P ≤ 0.01. (G) Kinetic analysis of M700V-EZH2 mutant on unmodified or H3K36cme2/3 12mer array substrates. Error bars represent SEM (n = 3). Table 1 presents associated kinetic values. (H) WT or disease-associated mutant EZH2-containing PRC2 HMT activity on H3K36cme3 mononucleosomes, normalized to each enzyme’s activity on H3K36cme0 mononucleosome substrates (dotted line). Error bars represent SEM (n = 3). *P ≤ 0.05; ns, not significant.