Significance
Ebolaviruses belong to the group of nonsegmented negative strand (NNS) RNA viruses. Most members of the Ebolavirus genus cause severe disease in humans. Here, we report that ebolaviruses have evolved a genome replication mechanism that distinguishes them from the other NNS RNA viruses. The ebolavirus replication strategy includes the addition of a nontemplated 3′ nucleotide at the genome ends which may aid in avoiding antiviral recognition. Our data highlight the need for more research on NNS RNA virus replication strategies to reveal commonalities between virus families as well as unique features. Dissecting the replication mechanisms of NNS RNA viruses, including ebolaviruses, paves the way for the development of broad-spectrum antivirals that target the viral replication complexes, thereby preventing virus amplification.
Keywords: Ebola virus replication, Ebola virus genome ends, variable 3′ genome ends, ebolavirus replication initiation, nonsegmented negative strand RNA virus replication
Abstract
Most nonsegmented negative strand (NNS) RNA virus genomes have complementary 3′ and 5′ terminal nucleotides because the promoters at the 3′ ends of the genomes and antigenomes are almost identical to each other. However, according to published sequences, both ends of ebolavirus genomes show a high degree of variability, and the 3′ and 5′ terminal nucleotides are not complementary. If correct, this would distinguish the ebolaviruses from other NNS RNA viruses. Therefore, we investigated the terminal genomic and antigenomic nucleotides of three different ebolavirus species, Ebola (EBOV), Sudan, and Reston viruses. Whereas the 5′ ends of ebolavirus RNAs are highly conserved with the sequence ACAGG-5′, the 3′ termini are variable and are typically 3′-GCCUGU, ACCUGU, or CCUGU. A small fraction of analyzed RNAs had extended 3′ ends. The majority of 3′ terminal sequences are consistent with a mechanism of nucleotide addition by hairpin formation and back-priming. Using single-round replicating EBOV minigenomes, we investigated the effect of the 3′ terminal nucleotide on viral replication and found that the EBOV polymerase initiates replication opposite the 3′-CCUGU motif regardless of the identity of the 3′ terminal nucleotide(s) and of the position of this motif relative to the 3′ end. Deletion or mutation of the first residue of the 3′-CCUGU motif completely abolished replication initiation, suggesting a crucial role of this nucleotide in directing initiation. Together, our data show that ebolaviruses have evolved a unique replication strategy among NNS RNA viruses resulting in 3′ overhangs. This could be a mechanism to avoid antiviral recognition.
Ebolaviruses belong to the filovirus family and have the potential to cause severe disease in humans with high case fatality rates. The Ebolavirus genus is subdivided into five distinct species, each defined by its best characterized member: Ebola virus (EBOV), Sudan virus (SUDV), Reston virus (RESTV), Tai Forest virus, and Bundibugyo virus. Ebolaviruses have nonsegmented negative strand (NNS) RNA genomes and share common mechanisms for mRNA transcription and genome replication with other NNS RNA viruses (1). At the 3′ and 5′ extremities of the ebolavirus genomes are short noncoding regions, the 3′ leader (Le(−)) and the 5′ trailer (Tr(−)), which contain essential regulatory elements for genome replication and transcription (2, 3). Genome replication depends on a bipartite promoter, which consists of an element within the 3′ Le(−) region and a second element within the first gene (4). This promoter directs the viral polymerase to synthesize a full-length, complementary copy of the genome, the antigenome. The replication promoter of the antigenome resides in the complementary trailer region (Tr(+)) and directs the viral polymerase to begin synthesis of genomic RNA. Both the genome and antigenome RNAs are encapsidated as they are synthesized, allowing the polymerase to be highly processive during replication (reviewed in refs. 5–7).
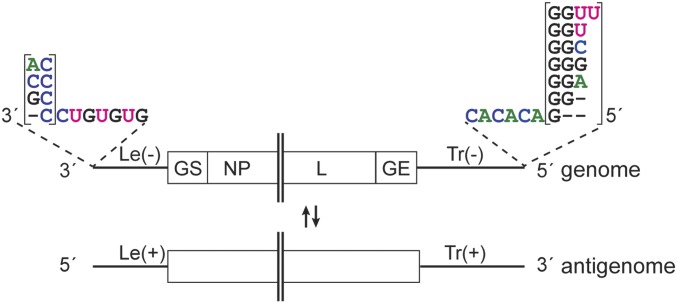
Studies on other NNS RNA viruses, including vesicular stomatitis virus (VSV), respiratory syncytial virus (RSV), and Nipah virus have shown that their polymerases initiate RNA replication opposite the first nucleotide of the promoter by a de novo, or primer-independent, initiation mechanism (8–10). However, although the overall genome structures of the ebolaviruses are similar to those of other NNS RNA viruses, and the active sites of their polymerases are well conserved (11, 12), ebolavirus genome ends are unconventional. Most NNS RNA viruses have almost identical promoters at the 3′ ends of their genome and antigenome RNAs, meaning that their ends show terminal complementarity, and these sequences are highly conserved within each virus species, reflecting their importance. In contrast, an early report by Kiley et al. (2), describing direct RNA sequencing of the 3′ termini of EBOV and SUDV genomes, determined that the 3′ terminal sequence was heterogeneous, with the terminal nucleotide being either a G or an A residue. This A/G heterogeneity is not reflected in more recent sequence analyses of EBOV or SUDV, or other ebolaviruses within the GenBank database, including the newly discovered Bombali virus (13). Instead, the majority of the deposited ebolavirus sequences starts with a 3′ terminal G residue, or the first nucleotide is not present (14). Likewise, the deposited ebolavirus genome sequences have variable 5′ ends that are not an exact copy of the 3′ termini, suggesting that the Le(−) and Tr(+) promoters differ with respect to the 3′ terminal nucleotides (Fig. 1 and SI Appendix, Table S1). These unusual promoter features suggest that ebolaviruses might have evolved a replication initiation mechanism that is different from other NNS RNA viruses.
Fig. 1.
Schematic diagram illustrating the variety of published ebolavirus 3′ and 5′ genome ends. The Le(−) region is followed by the gene start signal (GS) for the first gene, nucleoprotein (NP) gene. The Tr(−) region is preceded by the gene end signal (GE) for the L gene. Most of the viral genes are not shown. − symbols indicate the lack of a nucleotide. The complementary antigenome is depicted beneath the genome.
To gain a better understanding of the ebolavirus replication mechanism, we investigated the genome ends of ebolaviruses belonging to three different species, EBOV, SUDV, and RESTV. We found that they all have a variable 3′ terminal nucleotide on both the genome and antigenome RNAs. In contrast, the 5′ ends of the replicative RNAs are highly conserved among the different ebolaviruses and are one nucleotide shorter compared with the majority of the published 3′ ends. Consistent with these data, we show that in contrast to the other NNS RNA viruses studied to date, the ebolavirus polymerase is able to initiate RNA replication opposite position 2 of the template, and we present evidence that suggests the noncomplementary 3′ terminal sequences are added by a back-priming mechanism. Together, these data show that ebolaviruses have evolved a replication initiation strategy that is distinctive among NNS RNA viruses.
Results
Sequencing of the 3′ End of Ebolavirus RNAs Reveals Variability of the 3′ Terminal Nucleotide.
As noted above, the ebolavirus sequences deposited in GenBank show variability at the 5′ terminus of the Tr region, and suggest that the Tr(+) promoter at the 3′ end of the antigenome has a different sequence than the Le(−) promoter at the 3′ end of the genome (Fig. 1 and SI Appendix, Table S1). Terminal sequences of RNA molecules can be difficult to determine for technical reasons. For example, intragenic ligation of the genome ends does not allow to assign exactly which nucleotides lie at the 3′ versus 5′ terminus of the molecule. In addition, the identity of the 3′ terminal nucleotide and RNA structure affect RNA ligation efficiency which can lead to an artifactual bias toward particular sequences that are not representative of the population (15, 16). Therefore, to characterize the sequences at the 3′ ends of ebolavirus genome and antigenome RNAs, we utilized a 3′ rapid amplification of cDNA ends (3′ RACE) method in which the 3′ end of the RNA was tailed with a given NTP and poly(A) polymerase, followed by PCR amplification of ebolavirus-specific terminal sequences. The resulting PCR products were then sequenced. Studies have shown that the identity of the 3′ terminal nucleotide does not cause a significant bias in tailing efficiency, particularly if the RNA is tailed with CTP (17), and so this approach should yield a representative analysis of 3′ terminal sequences.
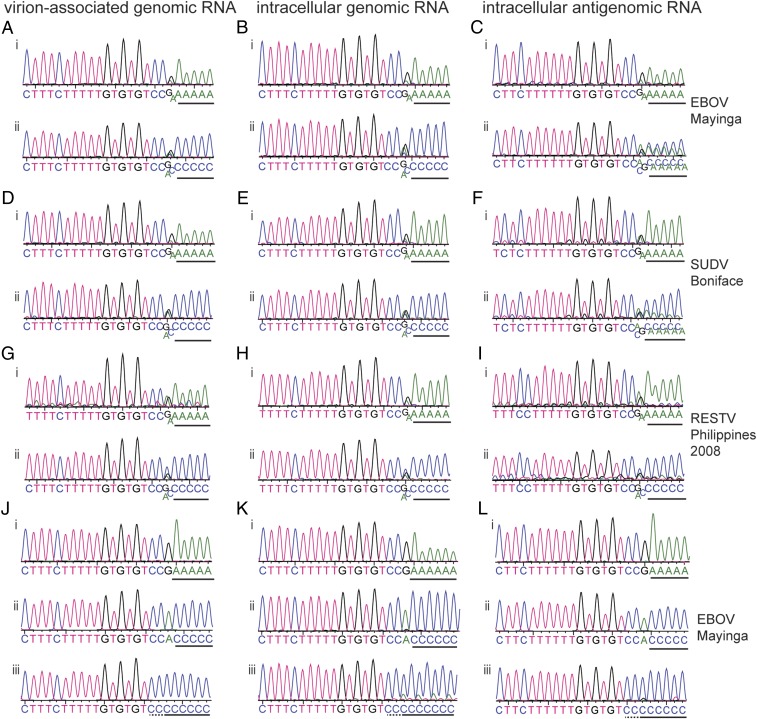
Sequence analysis of RNA purified from viral particles of the EBOV isolate Mayinga and tailed with ATP showed multiple peaks at the 3′ terminal nucleotide position of the traces, confirming heterogeneity at this position (Fig. 2 A, i) (2). Heterogeneity was also observed when the RNA was tailed with CTP (Fig. 2 A, ii). We used the same approach to sequence the 3′ terminus of intracellular genomic RNA, as it is possible there is a difference in the population of intracellular genome sequences compared with those packaged into viral particles. A similar heterogeneity to the virion-associated genomic RNA was observed (Fig. 2B). We also applied this technique to examine intracellular antigenomic RNA (Fig. 2C), and again saw heterogeneity at the 3′ terminal nucleotide. Similar results were obtained with the virus isolates SUDV Boniface (Fig. 2 D–F), RESTV Philippines 2008 (Fig. 2 G–I), EBOV Kikwit (SI Appendix, Fig. S1 A–C), and RESTV Pennsylvania (SI Appendix, Fig. S1 D and E). Thus, the 3′ terminal position of both genomic and antigenomic viral RNAs is heterogeneous.
Fig. 2.
Sequence analysis of the 3′ ends of ebolavirus RNAs. (A–I) Vero cells were infected with the indicated ebolavirus species and total cellular or virion-associated RNA was used for 3′ RACE and sequence analysis. The traces show sequences of the 3′ RACE PCR population obtained from virion-associated genomic RNA (A, D, and G) intracellular viral genomic RNA (B, E, and H), or intracellular viral antigenomic RNA (C, F, and I). In each case, i and ii show results from RNA tailed with ATP or CTP, respectively. (J–L) Representative traces of the most frequently observed sequences obtained from analysis of single cDNA clones of virion-associated (J), intracellular genomic viral RNA (K), or intracellular antigenomic viral RNA (L) isolated from Vero cells infected with EBOV Mayinga. Poly(A) (i) and poly(C) (ii and iii) sequences added during the 3′ RACE procedure are underlined with a black line. It should be noted that the polymeric sequence may include some virus-specific sequence that cannot be distinguished from the poly(A) or poly(C) tail (underlined with a dotted line).
Close examination of the sequence trace at the 3′ terminal position revealed that if RNA was tailed with ATP, the variable nucleotide was typically either a G or an A, but because the tail was composed of adenosine nucleotides, it was not possible to distinguish if the heterogeneity was due to the 3′ terminal nucleotide being an A, rather than G, or absent. In the sequence traces in which the RNA was tailed with CTP, the terminal nucleotide was either G, C, or A. Because a C residue was not present at the 3′ terminal position in the A-tailed sequences, we concluded that the majority of 3′ termini contained the sequences 3′ CCUGUGUG with either an additional 3′ A or G residue, or with no additional nucleotide. To confirm our finding, and to determine if there were other more minor populations, the 3′ RACE products corresponding to EBOV Mayinga RNA were inserted into a plasmid vector, and individual clones were isolated and sequenced. Traces representing the most frequently observed sequences are shown in Fig. 2 J–L, and a full list of sequences obtained including the respective clone frequencies is shown in SI Appendix, Table S2. Single clone sequencing confirmed that the 3′ terminal sequences of the viral genomes and antigenomes were typically either 3′ GCCUGUGUG, or 3′ ACCUGUGUG, with 3′ CCUGUGUG in some cases.
Other sequences were identified at a lower frequency (SI Appendix, Table S2). One clone contained a deletion at the 3′ end, another contained a U residue at the 3′ end, rather than an A or G residue, two clones contained two additional residues, 3′ UA or 3′ CA, and six clones, representing both genome and antigenome RNAs, contained longer additional sequence. Because the tailing reaction occurred before the reverse transcriptase and PCR steps in the 3′ RACE procedure, this indicates that the additional nucleotides were added by the EBOV polymerase rather than by reverse transcriptase. Of note, in the clones containing longer additional sequence, the added sequence was palindromic to the promoter sequence. This finding suggests that the nucleotide additions occurred by a back-priming event in which the 3′ end of the genome or antigenome folded back on the template and was extended by the EBOV polymerase for a few nucleotides (SI Appendix, Fig. S2 A–C). In other cases, the additional sequence was not an exact complement of the template, but showed some similarity. One possible explanation for this is that the 3′ end of the RNA folded and refolded more than once, leading to chimeric sequences being added (SI Appendix, Fig. S2 D–F). These data suggest that the 3′ termini of EBOV genome and antigenome RNAs have the capability to fold into secondary structures and be extended by the viral polymerase.
The EBOV Polymerase Initiates RNA Synthesis Opposite the First C Residue of the 3′ CCUGUG Motif.
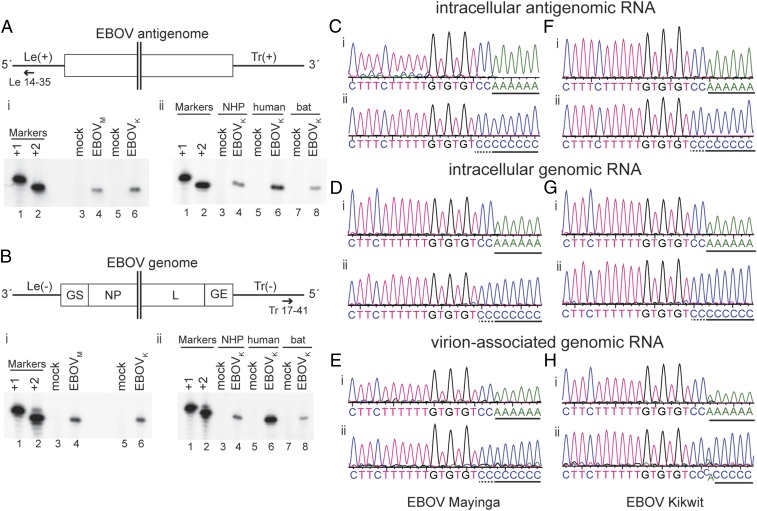
It is generally accepted that NNS RNA virus polymerases initiate genome replication opposite the 3′ terminal nucleotide of the template. Given that ebolavirus genomes and antigenomes are variable at the 3′ terminal position, this raises the question of where replication is initiated. To determine this, the 5′ end of EBOV antigenome RNA was mapped using primer extension analysis. Total intracellular RNA was isolated from Vero cells infected with EBOV isolates Mayinga or Kikwit and analyzed using primers that annealed near the 5′ of the antigenome (Fig. 3A). RNA from mock-infected Vero cells was used as control. The radiolabeled cDNA products were analyzed by denaturing gel electrophoresis alongside DNA oligonucleotide markers representing RNA initiated at position 1 or 2 of the template relative to the published 3′ terminal sequence of the Le(−) promoter. In the samples from EBOV-infected cells, a single band was detected, which migrated comparably to the marker representing initiation at position 2 (Fig. 3 A, i). These findings indicate that antigenome synthesis was initiated opposite the first C residue of the 3′ CCUGUGUG motif. To confirm this, the 5′ terminal sequence of the EBOV Mayinga antigenome was determined by performing 5′ RACE and sequence analysis of the PCR product (Fig. 3C). In contrast to the 3′ RACE products described above, there was no observable heterogeneity at the 5′ terminus of the antigenome RNA in the population analysis, and sequence analysis of single DNA clones revealed that in most cases, the 5′ terminal sequence was 5′ GGACAC consistent with initiation from the first C residue of the promoter. Only two clones with varying additional nucleotides were observed (SI Appendix, Table S2). Neither contained the repetitive sequences that were occasionally detected in the 3′ RACE analysis, but instead contained one or two additional nucleotides that could either have arisen due to the polymerase initiating opposite the 3′ nucleotide, rather than internally, or due to nucleotide addition by reverse transcriptase during cDNA synthesis (note that in contrast to 3′ RACE, during 5′ RACE, cDNA synthesis by reverse transcriptase precedes the tailing reaction, meaning that in this case, reverse transcriptase errors cannot be distinguished from the true RNA sequence). The 5′ end of viral genomic RNA was also examined and yielded similar results as the 5′ end of viral antigenomic RNA (Fig. 3 B, i, D and E). Similar results were obtained for EBOV Kikwit (Fig. 3 F–H), SUDV Boniface (SI Appendix, Fig. S3), RESTV Pennsylvania, and RESTV Philippines 2008 (SI Appendix, Fig. S4).
Fig. 3.
Mapping the 5′ end of EBOV RNAs by primer extension and sequence analysis. (A and B) Primer extension analysis of antigenome (A) and genome (B) RNAs. (A and B, Upper) Schematic diagram (not to scale) of the EBOV RNA that was analyzed showing the hybridization positions of the negative sense Le 14–35 (A) and positive sense Tr 17–41 (B) primers used for primer extension analysis. (A and B, Lower) Primer extension analysis. In each case, i shows analysis of RNA isolated from Vero cells infected with EBOV Mayinga (EBOVM) or Kikwit (EBOVK), and ii shows analysis of RNA isolated from EBOV Kikwit-infected Vero (nonhuman primate, NHP), Huh7 (human), and R05T (bat) cells. In each panel, [γ-32P]ATP end-labeled DNA oligonucleotides corresponding in length and sequence to cDNA representing initiation from positions +1 and +2, relative to the published EBOV sequences, were used as markers (lanes 1 and 2). (C–H) Vero cells were infected with EBOV Mayinga (C–E) or Kikwit (F–H) and total cellular or virion-associated RNA was used for RACE analysis. The traces show sequences of the 5′ RACE PCR population obtained from the 5′ ends of intracellular antigenomic viral RNA (C and F), intracellular genomic viral RNA (D and G), or virion-associated genomic RNA (E and H). The cDNA was tailed with dATP (i) or dCTP (ii). The black line below the sequence traces indicates poly(A) or poly(C) tail sequences that had been added to the virus-specific sequence during RACE. This may include some virus-specific sequence that cannot be distinguished from the poly(A) or poly(C) tail. The first two C residues that belong to the viral sequence are underlined with a dotted line in the poly(C)-tailed sequence traces.
In contrast to humans and nonhuman primates who are highly susceptible to EBOV infection and develop symptoms of severe disease (18), Rousettus aegyptiacus fruit bats are not susceptible, although cell lines derived from these bats are permissive to EBOV infection (19–21). To explore if EBOV replication initiation varies depending on the host cell, we infected cell lines derived from nonhuman primates (Vero cells), humans (Huh7 cells), and R. aegyptiacus fruit bats (R05T cells) with EBOV and performed primer extension analysis using antigenomic and genomic RNA. In all cell lines, the cDNA products migrated with the +2 marker, confirming replication initiation at position +2 (Fig. 3 A, ii and B, ii). In conclusion, our data indicate that RNA synthesis of all tested ebolaviruses is initiated opposite the first C residue of the 3′ CCUGUG motif, at position 2 relative to the published sequence. The +2 replication initiation does not seem to be dependent on the host cell.
The EBOV Polymerase Initiates RNA Synthesis Opposite 3′ CCUGUG, Regardless of Identity of the 3′ Terminal Nucleotide.
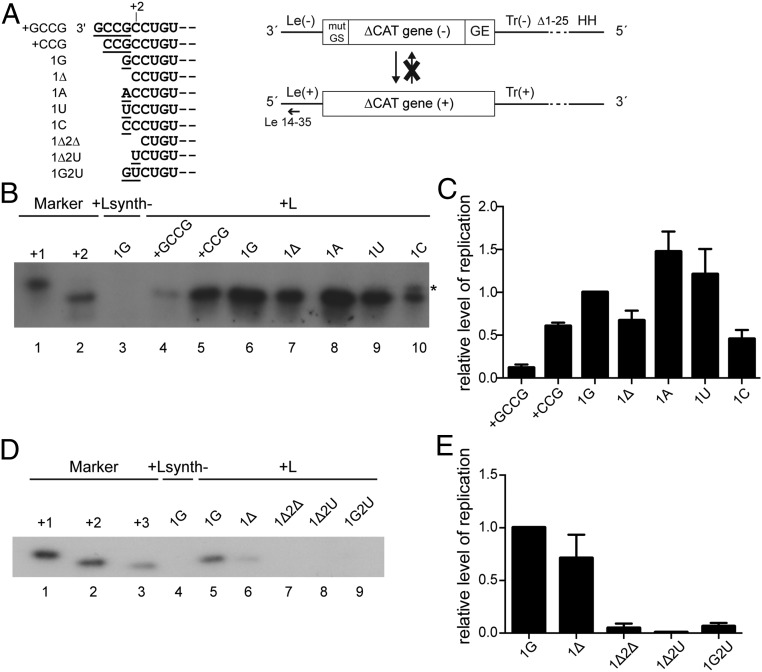
The data presented above show that the templates for replication are heterogeneous at their 3′ terminal nucleotide, but that replication products are initiated opposite the first nucleotide of the 3′ CCUGUG motif (the second nucleotide of the genome relative to most previously published sequences). However, this sequence analysis did not show if all genome and antigenome sequences were functional templates, or if only a subset could be used. For example, it was possible that only the templates lacking the 3′ terminal A or G (i.e., containing 3′ CCUGUG) were functional. Therefore, to determine what constitutes a functional template, EBOV minigenomes were generated which either did not contain an additional nucleotide (1Δ), or contained the additional 3′ terminal A or G residue. We also tested minigenomes containing an additional 3′ terminal U or C residue, as well as minigenomes that contained additional 3′ nucleotides (+GCCG, +CCG), to test the accuracy of initiation. To ensure that the 3′ termini of the minigenomes were not modified during multiple replication cycles, the minigenomes were restricted to the antigenome synthesis step of replication by deleting the 5′ terminal 25 nucleotides of the Tr region (Fig. 4A). In addition, the EBOV-specific gene start signal (GS), which is required for mRNA transcription, was mutated to suppress minigenome transcription. BSR T7/5 cells were transfected with the respective minigenome DNA along with expression plasmids encoding the EBOV proteins required for replication (NP, VP35, VP30, and L), and the RNA products generated from the minigenome templates were analyzed by primer extension (Fig. 4 B–E) and by Northern blot hybridization to complement the primer extension analysis (SI Appendix, Fig. S5C). All minigenomes with substitutions at position 1, a deletion at position 1, or additional nucleotides added to the 3′ terminus were functional templates for replication, although the minigenome containing an additional GCCG sequence at the 3′ end yielded antigenome RNA at such a low level that it could not be detected by Northern blot (SI Appendix, Fig. S5C, lane 2) and was only barely detectable by primer extension analysis (Fig. 4 B, lane 4 and C). RNA from the remaining mutant minigenomes in Fig. 4B could be detected by Northern blot analysis, confirming the results by primer extension (SI Appendix, Fig. S5C). Interestingly, the primer extension analysis showed that regardless of the minigenome 3′ terminal sequence, the EBOV polymerase initiated RNA synthesis at the same position as observed in viral infection, i.e., opposite the first C residue of the CCUGUG motif (Fig. 4B). Thus, it initiated RNA replication opposite position +1 in the 1Δ mutant, opposite position +2 in mutants 1G, 1A, 1U, and 1C, and opposite positions +4 and +5, respectively, in mutants +CCG and +GCCG (Fig. 4B). These data suggest that the EBOV polymerase locates its binding site within the promoter sequence and initiates RNA synthesis accurately, regardless of the identity of the 3′ terminal nucleotide(s) and of the position of the promoter sequence relative to the 3′ end. The only exception was mutant 1C, which presented with a double band in the primer extension analysis (Fig. 4B, lane 10, asterisk). This suggests that, in this case, on some occasions, the EBOV polymerase initiated opposite the additional C residue at the first nucleotide of the template.
Fig. 4.
Analysis of replicated RNA produced from EBOV minigenomes with different 3′ ends. (A) Schematic diagram (not to scale) showing the different 3′ terminal sequences that were tested (Left) and the organization of the single-cycle replicating, nontranscribing EBOV minigenome and the antiminigenome product (Right). The terminal 25 nucleotides of the trailer were deleted (Δ1–25). The trailer is flanked by an inactive hammerhead ribozyme (HH). The annealing position of the negative sense primer used for primer extension is indicated; mut GS, mutated gene start signal; GE, gene end signal. (B) Primer extension analysis of antiminigenome RNA from minigenomes with +GCCG, +CCG, 1G, 1Δ, 1A, 1U, and 1C 3′ ends using a negative sense primer that annealed within the Le(+) region. The markers are [γ-32P]ATP end-labeled DNA oligonucleotides corresponding in length and sequence to cDNA representing initiation from positions +1 and +2, relative to published sequence. As a negative control, L was replaced with the enzymatically inactive Lsynth- mutant (52). A representative result of three independent experiments is shown. (C) Quantification of primer extension products shown in B. The data are normalized to the level of product generated by the 1G minigenome after subtraction of the Lsynth- negative control. Shown are the mean and SE of three independent experiments. (D) Primer extension analysis of antiminigenome RNA from minigenomes with 1G, 1Δ, 1Δ2Δ, 1Δ2U, and 1G2U 3′ ends using a negative sense primer that annealed within the Le(+) region. The markers are [γ-32P]ATP end-labeled DNA oligonucleotides corresponding in length and sequence to cDNA representing initiation from positions +1, +2, and +3 relative to published sequence. As a negative control, L was replaced with the enzymatically inactive Lsynth- mutant (52). A representative result of three independent experiments is shown. (E) Quantification of primer extension products shown in D. The data are normalized to the level of product generated by the 1G minigenome after subtraction of the Lsynth- negative control. Shown are the mean and SE of three independent experiments.
The EBOV Polymerase Does Not Initiate RNA Synthesis Opposite the 3′ CCUGUG Motif if the First Nucleotide of This Motif Is Missing or Substituted.
The data presented above show that EBOV replication products are initiated opposite the first nucleotide of the 3′ CCUGUG motif, regardless of the presence or identity of additional 3′ nucleotides. Interestingly, the genome ends of the closely related Marburg virus (MARV) begin with a similar 3′ UCUGUG motif, with the only differences between the EBOV and MARV genome ends being the terminal nucleotide of this motif (C versus U) and the lack of an additional, nontemplated nucleotide at the 3′ end of the MARV genome (2). To determine the role of the first C residue of the 3′ CCUGUG motif in EBOV replication, single-round replicating EBOV minigenomes were generated which lacked both the 3′ terminal, nontemplated nucleotide and the first C residue of the CCUGU motif (1Δ2Δ), or had this C residue mutated to U in the absence of an additional nucleotide (1Δ2U), or had this C residue mutated to U in combination with an additional terminal G residue (1G2U) (Fig. 4A). Intriguingly, none of these minigenomes were accepted as templates for replication initiation, as shown by primer extension analysis (Fig. 4 D, lanes 7–9 and E).
To confirm the key importance of the first C residue of the CCUGUG motif in the EBOV replication cycle, replication- and transcription-competent minigenomes in which the C residue was mutated to U (1G2U and 1Δ2U) were generated and tested. The readout in this assay was minigenome-derived reporter gene expression (SI Appendix, Fig. S5D). The mutant minigenomes produced only background levels of reporter gene activity in contrast to the strong activity seen with the 1G minigenome which was used as a positive control (SI Appendix, Fig. S5E). In combination with the primer extension data, these results show that the first C nucleotide of the 3′ CCUGUG motif is absolutely required for EBOV genome replication. Of note, the second nucleotide in this motif is a C residue, too. The fact that this is not sufficient to allow replication shows that a 3′ terminal C residue alone is not sufficient to initiate replication; but rather that this nucleotide functions in combination with other nucleotides to direct initiation.
Discussion
The data we present here show that the 3′ ends of the EBOV genomes and antigenomes contain a variable nucleotide, and that the polymerase initiates at the first C residue of the template, typically at position +2 (Fig. 5). The identity and presence of this nucleotide are crucial for EBOV genome replication. These conclusions are drawn from a number of experiments, including sequence analysis of the 3′ ends of viral genome and antigenome RNA, both of populations and individual clones, primer extension, minigenome activity, and sequence analysis of the 5′ end of the RNAs. All data obtained in this study were consistent, regardless of the technique used. Further, these findings are applicable across the ebolaviruses. Importantly, our results confirm previous findings by Kiley et al. (2) deduced from chemical RNA sequence analysis that the 3′ terminal nucleotide of EBOV and SUDV genomes is either a G or an A residue. A study of EBOV Makona isolates had also found heterogeneity at the 3′ termini, reporting that most RNAs contained a 3′ G residue, with a minor fraction that was one nucleotide shorter. However, in that study a terminal A residue was not reported (14). Our data, and that of Kiley et al. (2) , differ from most of sequences deposited in GenBank, in which the 3′ terminal residue of the ebolavirus genome is shown as a G residue or not present, and the 5′ terminus is variable. We believe that the discrepancy between the published ebolavirus genome ends is likely due to technical reasons, such as the inherent bias of RNA ligation, PCR artifacts, or sequencing of individual clones, rather than the RNA population, and the fact that it is not possible to categorically assign the 3′ and 5′ terminal sequences when the RNA is circularized before sequencing.
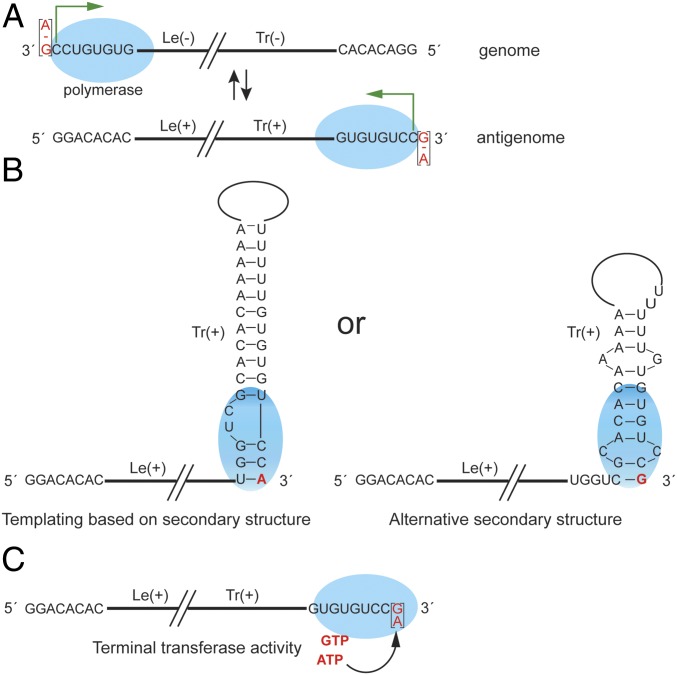
Fig. 5.
Model for RNA synthesis initiation and 3′ terminal nucleotide addition by the ebolavirus polymerase. (A) Scheme showing the site of initiation at the Le(−) promoter of the genome and at the Tr(+) promoter of the antigenome. The ebolavirus polymerase is indicated by a blue ellipse and the initiation site by a green arrow. The experimentally determined 3′ terminal nucleotide is highlighted in red. − symbols indicate the lack of a nucleotide. (B) Schematic diagram showing how addition of a G or A residue onto the 3′ end of antigenome RNA may occur by templating of the 3′ terminal nucleotide using secondary structure formations at the 3′ end of the RNA. (C) Schematic diagram showing addition of a 3′ terminal purine nucleotide by terminal nucleotidyl transferase activity of the ebolavirus polymerase or a cellular terminal transferase.
Analysis of the 5′ termini of ebolavirus genome and antigenome RNAs suggested that the polymerase could initiate RNA replication internally on the template, at the first C of the sequence 3′ NCCUGUGUG (Fig. 3). However, there were two possible alternative explanations. First, it was possible that the polymerase initiated opposite the 3′ N residue, and that the 5′ terminal nucleotide of the replication product was cleaved. A similar mechanism has been shown for the NNS RNA virus, Borna disease virus (22). However, other data indicate that this is not the case. If the RNA were cleaved, it would contain a 5′ monophosphate rather than triphosphate moiety, but previous studies have shown that EBOV genome RNA stimulates retinoic acid-inducible gene I (RIG-I) activation, an activity that can be ablated by treating the RNA with phosphatase (23). Thus, the 5′ end of EBOV genome RNA contains a 5′ triphosphate, excluding the possibility of nucleotide removal by cleavage. The second possible explanation was that genomic and antigenomic viral RNAs containing an additional 3′ terminal nucleotide were defective, and only RNA templates with the sequence 3′ CCUGUGUG were viable. However, experiments with the minigenome system confirmed that templates with any nucleotide at the 3′ N position were recognized by the polymerase. The polymerase initiated opposite the first C residue of the 3′ CCUGUGUG motif, even if the template contained up to four additional nucleotides at the 3′ end (Fig. 4). Thus, the ebolavirus polymerase can initiate internally on a template. To our knowledge, this is a previously unreported instance of NNS RNA viruses initiating RNA replication internally on the template as part of their normal replication cycle.
Our data also suggest that the promoter sequence plays a key role in determining the site of RNA synthesis initiation, as opposed to it being determined by the 3′ terminus of the RNA template. This mechanism of initiation site selection has also been described for other NNS RNA viruses: while Sendai virus and RSV polymerases normally only initiate RNA replication opposite the 3′ terminal nucleotide of the template, they are both capable of initiating RNA replication opposite an internal promoter sequence when presented with an artificial template containing additional 3′ nucleotides (24–26). Thus, the ability of the ebolavirus polymerase to initiate at an internal site is not unique, but the fact that replication is initiated at an internal site as part of the normal replication cycle is distinctive among NNS RNA viruses. While the promoter appears to play a dominant role in positioning the polymerase opposite the initiation site, the presence of the C residue at the initiation site is also important as a minigenome template containing an additional 3′ terminal C residue showed a low level of initiation from the first (3′ terminal) C residue, as well as from the typical initiation site (Fig. 4B, lane 10). Given that the ebolavirus gene start signals begin with a C residue (27, 28), this could reflect a preference of the polymerase for initiating with GTP. A recent study of RSV initiation showed that its polymerase has a template-independent affinity for the initiating NTPs and that this plays a role in initiation site selection (29). Thus, it is possible that ebolavirus polymerases preferentially bind GTP due to template-independent affinity, and that this affinity combined with positioning by the promoter helps guide initiation at the correct site.
In contrast to the flexibility seen with the EBOV replication complex with regards to accommodating extra, nontemplated 3′ terminal nucleotides, data presented here indicate that both the identity and the presence of the first C residue of the 3′ CCUGUGUG motif are crucial for EBOV replication (Fig. 4D and SI Appendix, Fig. S5E). The homologous region in the MARV leader starts with a U residue (3′ UCUGUGUG) (2). Despite the similarities of the EBOV and MARV leader regions, EBOV is not able to initiate replication if the first nucleotide of the 3′ CCUGUGUG motif is mutated to a U. Intriguingly, this strict requirement for a C residue at the first position of the CCUGUGUG motif has also been seen with Lloviu virus, a new member of the filovirus family, but not with MARV or the recently discovered MARV-like Měnglà virus, whose replication complexes are each able to replicate the EBOV leader (30–33). This divergence in terms of nucleotide specificity may indicate either a divergent mechanism for the initiation of replication between these two branches of the filovirus family tree or, at the very least, a relaxed specificity for the MARV/Měnglà replication complex.
Given that the ebolavirus polymerases initiate at position +2 of the template, the 3′ terminal nucleotide of the genome and antigenome RNAs is not templated by the complementary strand. This leads to the question of how the 3′ terminal nucleotides are added to the genomic and antigenomic RNAs. Two possible models exist: 3′ extension by back-priming or nucleotide addition by terminal transferase activity (Fig. 5). The back-priming model postulates that the 3′ end of the RNA folds into a hairpin structure, allowing templated addition of the 3′ terminal nucleotides. While the dogma in the NNS RNA field is that encapsidation of replicative RNA by nucleoprotein prevents secondary structure formation, 3′ extension of viral replicative RNA by back-priming has been shown to occur in Borna disease virus and on some antigenome RNAs in RSV-infected cells (9, 34), indicating that encapsidation of RNA at 3′ ends of the replicative RNA is sometimes incomplete or delayed. The data obtained in this study support this model, as some 3′ termini contained palindromic sequences, which is consistent with the RNA having been folded into a hairpin and extended (SI Appendix, Fig. S2). In addition, the EBOV RNA has the potential to form more extensive and stable secondary structures (3, 4, 35), which would allow templating of either an A or G residue, depending on the secondary structure formed (Fig. 5B). Regarding the possibility of terminal transferase activity, it has been observed that several RNA-dependent RNA polymerases possess terminal transferase activity (36–40) and it is possible that ebolavirus polymerases share this property. Given that the additional 3′ terminal nucleotide is typically an A or G residue, if this model is correct, it suggests that the polymerase has a preference for performing terminal transferase addition with purine nucleotides. The identity of an extra nucleotide to the 3′ end of the genome did not appear to affect sorting of genome RNAs into virions (Fig. 2 and SI Appendix, Fig. S1) and was not required for replication to occur (Fig. 4), but it did lead to a marginal increase in replication efficiency. Thus, it is possible that the presence of an additional nucleotide benefits the virus by providing enhanced stability to the polymerase complex during initiation.
In addition to direct effects on viral genome replication, ebolaviruses might contain an additional 3′ nucleotide to reduce detection by RIG-I and RIG-I-like receptors whose binding to dsRNA is impaired by 3′ overhangs (41, 42). Interestingly, EBOV VP35, a viral dsRNA binding protein involved in viral replication and suppression of IFN induction, also binds dsRNA with 3′ overhangs with reduced affinity compared with blunt and 5′ overhang dsRNA, although if or how this would benefit the virus remains unclear (43). It has been shown for other negative sense RNA viruses that they use unconventional replication initiation mechanisms to avoid antiviral responses. This includes Borna disease virus, a NNS RNA virus, and Hantaan virus, a segmented negative strand RNA virus, which both use distinct replication initiation mechanisms to remove RIG-I stimulating 5′ triphosphates from their genome ends (22, 23, 44). Similar to ebolaviruses, Tacaribe virus, a segmented negative strand RNA virus that belongs to the arenavirus family, initiates RNA synthesis at position +2 on the template. However, in this case, the internally initiated RNA is used in a prime-realign mechanism which results in a one-nucleotide 5′ overhang that interferes with RIG-I recognition (45, 46). In contrast, our data for ebolaviruses suggest prime-realign does not occur and a 3′ overhang is generated.
In summary, both the 3′ and 5′ ends of ebolavirus replicative RNAs are formed by unusual mechanisms, with the 5′ terminus being generated by internal initiation, and the 3′ terminus being generated by nucleotide addition by back-priming and/or terminal transferase activity with high frequency. These findings reveal an unappreciated variety between NNS RNA viruses.
Materials and Methods
Cell Lines and Viruses.
African green monkey kidney cells (Vero; ATCC CRL-1586), human hepatocellular carcinoma cells (Huh7), and Egyptian fruit bat (R. aegyptiacus) fibroblast cells (R05T) (47) were used for infection with ebolaviruses. Human embryo kidney cells (HEK 293T; ATCC CRL-3216) were used for the minigenome studies with replicating minigenomes. Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FBS (FBS), penicillin (50 units/mL), and streptomycin (50 mg/mL). BSR T7/5 cells constitutively expressing the T7 RNA polymerase and cells were used for the minigenome studies with single-round replicating minigenomes and maintained as described in ref. 48. EBOV human isolates Kikwit (GenBank: KR867676) and Mayinga (GenBank: AF086833), RESTV macaque isolate Pennsylvania (GenBank: AY769362) and porcine isolate Philippines 2008 (GenBank: FJ621583), and SUDV human isolate Boniface (GenBank: FJ968794) were propagated in Vero cells as previously described (49). All work with infectious EBOV, RESTV, and SUDV was performed in the biosafety level 4 facility of the Integrated Research Facility, Rocky Mountain Laboratories, Division of Intramural Research, NIAID, NIH, Hamilton, MT following standard operating procedures approved by the Institutional Biosafety Committee (IBC).
RNA Isolation from Infected Cells.
Cells seeded at 50–70% confluency in T75 or T150 flasks were infected with EBOV, RESTV, or SUDV at a multiplicity of infection (MOI) of 0.01 for EBOV and 0.001 for RESTV and SUDV, or mock infected. When the infected cells showed a significant cytopathic effect or 15 d post infection, supernatants containing viral particles were clarified by low-speed centrifugation (5,000 × g) before polyethylene glycol (PEG) precipitation of the viral particles (23). Pelleted viral particles, cell pellets, and monolayers of infected cells were lysed using TRIzol reagent (Invitrogen) according to the BSL-4 standard operating procedures for RNA purification approved by the IBC of the NIAID Rocky Mountain Laboratories (50). Following isopropanol precipitation, RNA was purified by an additional acid-phenol:chloroform extraction step that aids in the removal of DNA followed by ethanol precipitation. Isolated RNA was used for analysis by primer extension or 3′ and 5′ RACE followed by sequencing (51).
Primer Extension Analysis.
RNA 5′ ends were analyzed by reverse transcription of cDNAs with end-labeled PAGE-purified primers. Two micrograms of total cellular RNA from infected or transfected cells (or normalized to same amount of lowest RNA sample in the case of transfected cells) was combined with 0.2 pmol of a 32P end-labeled primer (SI Appendix, Table S3). The mixture was heated to 80 °C for 3 min, cooled on ice, and the resulting RNA-DNA hybrid was reverse transcribed using Sensiscript reverse transcriptase (Qiagen), according to the manufacturer’s instructions. End-labeled oligonucleotides corresponding to initiation from position +1 and +2 were used as markers (SI Appendix, Table S3). Samples were subjected to electrophoresis on 6 or 8% polyacrylamide gels containing 7 M urea and radiolabeled products were detected by autoradiography and quantified using phosphorimage analysis.
RACE Analysis.
For 3′ RACE, 2 µg of total intracellular RNA (viral genome and antigenome) or 500 ng of virion RNA was tailed with ATP or CTP using Escherichia coli poly(A) polymerase (NEB), followed by heat inactivation of the enzyme according to the manufacturer’s instructions. First-strand cDNA synthesis was performed using primers that annealed to the poly(A) or poly(C) tail, and Sensiscript (Qiagen) reverse transcriptase was used to reverse transcribe the tailed RNA according to the manufacturer’s instructions. The resulting cDNA was amplified by PCR using a sequence-specific primer and a primer that annealed to the poly(A) or poly(C) tail.
For 5′ RACE, 2 µg of total intracellular RNA (viral genome and antigenome) or 500 ng viral RNA (genome) was annealed to a virus-specific primer and reverse transcribed using Sensiscript reverse transcriptase, according to the manufacturer’s instructions. Following purification, the cDNA was tailed with dCTP or dATP using terminal transferase (NEB). The tailed cDNA was PCR amplified using a nested virus-specific primer and a primer that annealed specifically with the dATP or the dCTP tail. All PCRs were performed using Platinum Taq PCR (Invitrogen). PCR products were gel purified and either sequenced directly or cloned into the pCR2.1 vector (TA TOPO cloning kit; Life Technologies) for sequencing of individual cDNA clones.
All sequencing traces were analyzed using the SeqMan, DNASTAR Lasergene software. Segments of the sequencing traces were copied from SeqMan and inserted in the figures.
Cloning of Minigenomes.
Construction of nonreplicating minigenomes.
Minigenome mutants were generated based on the negative sense EBOV minigenome 3E5E (30). To optimize antiminigenome detection in the Northern blot analysis, the minigenome was shortened from its original length of 1879 nucleotides to 505 nucleotides. In the shortened minigenome, the trailer region contains the 129 terminal nucleotides of the EBOV Mayinga genome sequence and is flanked by a hammerhead ribozyme (52). The leader region is identical to minigenome 3E5E and is fused to a truncated version of the chloramphenicol acetyl transferase reporter gene (ΔCAT). Precise 3′ ends are generated by hepatitis delta virus ribozyme activity (30). To construct single-cycle replication minigenomes, the terminal 25 nucleotides of the trailer were removed (53). This had two effects, first it deleted the promoter at the 3′ end of the antigenome, preventing production of newly synthesized genome RNA; it also inactivated a hammerhead ribozyme positioned between the Tr region and the T7 promoter, so that the minigenome contained an irrelevant 5′ addition that did not resemble the EBOV promoter. To destroy the EBOV-specific GS signal required to initiate transcription (28), uridine residues 57, 60, and 61, which are located within the conserved GS signal, were substituted with adenosine (numbers refer to EBOV Mayinga sequence, GenBank AF086833). The various 3′ terminal nucleotides of the minigenome mutants are depicted in Fig. 4.
Construction of replicating EBOV minigenomes.
Minigenome mutants were generated based on the negative sense EBOV minigenome 3E5E-Luc (54). The 1G2U and 1Δ2U mutants are depicted in Fig. 4A.
Nonreplicating, Nontranscribing Minigenome Assays.
BSR T7/5 cells were seeded at 4 × 105 cells per well and grown overnight to ∼70% confluence in 6-well plates and transfected with 0.1 µg pTM1-LEBOV, 0.5 µg pTM1-NPEBOV, 0.5 µg pTM1-VP35EBOV, 0.1 µg pTM1-VP30EBOV (30), and 1.5 µg of the respective minigenome construct per well using Lipofectamine LTX with PLUS Reagent (ThermoFisher Scientific), following the manufacturer’s recommendations. All transfection mixtures were adjusted to the same total amount of DNA using pTM1 plasmid. As negative control, plasmid pTM1-LEBOV was replaced with the appropriate amount of pTM1-Lsynth- encoding a replication-deficient polymerase with a substitution in the catalytic site (52). Total cellular RNA was isolated at 2 d post transfection using TRIzol reagent as described above and used for primer extension and Northern blot analysis.
Replication- and Transcription-Competent Minigenome Assays.
The HEK 293T cells were seeded at 2 × 105 cells per well and grown overnight to ∼70% confluence in 12-well plates and transfected with 0.5 µg pCAGGS-LEBOV, 0.25 µg pCAGGS-NPEBOV, 0.25 µg pCAGGS-VP35EBOV, 0.05 µg pCAGGS-VP30EBOV (30), 1.0 µg pCAGGS-T7, 0.05 µg pTM1-β-gal, and 1.0 µg of the respective minigenome construct per well using TransIT-LT1 Transfection Reagent (Mirus Bio LLC), following the manufacturer’s recommendations. All transfection mixtures were adjusted to the same total amount of DNA using pCAGGS plasmid. As negative control, plasmid pCAGGS-LEBOV was replaced with the appropriate amount of pCAGGS-Lsynth- encoding a replication-deficient polymerase with a substitution in the catalytic site (52). Cell lysates were generated at 2 d post transfection using Cell Extraction Buffer (Thermo Fisher Scientific) and analyzed by luciferase and β-galactosidase assays.
Northern Blot Analysis.
To detect full-length replication products generated from minigenomes, RNA samples were subjected to electrophoresis in 1.5% agarose-formaldehyde gels in MOPS buffer and subjected to Northern blot analysis as previously described (55). In each experiment, the levels of input minigenome RNA were determined by probing Northern blots with a positive sense 32P-labeled CAT-specific riboprobe. Positive sense antiminigenome RNA was determined by probing Northern blots with a negative sense 32P-labeled CAT-specific riboprobe. DynaMarker Prestain Marker for RNA High was used along RNA from nonreplicating, nontranscribing minigenome assay to size the antiminigenome product (SI Appendix, Fig. S5B).
Luciferase and β–Galactosidase Assays.
To determine minigenome activity, cell lysates were diluted 1:100 in 1× Reaction Lysis Buffer (Promega). Diluted lysates (50 µL) were mixed with 50 µL of Firefly Luciferase Reagent (Promega), and luciferase activity was measured using a BMG Labtech Omega luminometer. To account for potential differences in transfection efficiency, luciferase values were normalized to β-galactosidase values. Undiluted cell lysates (50 µL) were mixed with 50 µL of 2× Assay Buffer (Promega) and incubated for 30 min at 37 °C. Reaction was terminated by adding 150 µL of 1 M sodium carbonate (Promega). β-galactosidase values were measured on a Tecan Spark microplate reader at 420 nm and normalized to a standard curve generated with the β-galactosidase provided by Promega.
Supplementary Material
Acknowledgments
We thank H. Feldmann (LV, NIAID, NIH) and members of the Laboratory of Virology for training L.R.D. in BSL-4 work and for help with BSL-4 experiments at the NIAID/NIH Rocky Mountain Laboratories, Hamilton, MT. We thank U.J. Buchholz, NIAID/NIH, Bethesda, MD for providing BSR T7/5 cells; J. Alonso, Texas Biomedical Research Institute, San Antonio, TX for sharing Huh7 cells; I. Jordan, ProBioGen AG, Berlin, Germany for providing R05T cells; T. Takimoto, St. Jude Children’s Research Hospital, Memphis, TN, and Y. Kawaoka, University of Wisconsin, Madison, WI for providing the pCAGGS-T7 plasmid; and J.R. Pacheco, Boston University for excellent technical assistance. This work was funded by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Awards R03 AI114293 (to E.M.), R21 AI126457 (to E.M.), R01 AI133486 (to R.F. and E.M.), and R01 AI113321 (to R.F.). The work was supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (to H.E.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815745116/-/DCSupplemental.
References
- 1.Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 2.Kiley MP, Wilusz J, McCormick JB, Keene JD. Conservation of the 3′ terminal nucleotide sequences of Ebola and Marburg virus. Virology. 1986;149:251–254. doi: 10.1016/0042-6822(86)90127-3. [DOI] [PubMed] [Google Scholar]
- 3.Volchkov VE, et al. Characterization of the L gene and 5′ trailer region of Ebola virus. J Gen Virol. 1999;80:355–362. doi: 10.1099/0022-1317-80-2-355. [DOI] [PubMed] [Google Scholar]
- 4.Weik M, Enterlein S, Schlenz K, Mühlberger E. The Ebola virus genomic replication promoter is bipartite and follows the rule of six. J Virol. 2005;79:10660–10671. doi: 10.1128/JVI.79.16.10660-10671.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolesnikova L, Nanbo A, Becker S, Kawaoka Y. Inside the cell: Assembly of filoviruses. Curr Top Microbiol Immunol. 2017;411:353–380. doi: 10.1007/82_2017_15. [DOI] [PubMed] [Google Scholar]
- 6.Kirchdoerfer RN, Wasserman H, Amarasinghe GK, Saphire EO. Filovirus structural biology: The molecules in the machine. Curr Top Microbiol Immunol. 2017;411:381–417. doi: 10.1007/82_2017_16. [DOI] [PubMed] [Google Scholar]
- 7.Brauburger K, Deflubé LR, Mühlberger E. Filovirus transcription and replication. In: Pattnaik AK, Whitt MA, editors. Biology and Pathogenesis of Rhabdo- and Filoviruses. World Scientific Publishing Co. Pte. Ltd.; Singapore: 2015. pp. 515–555. [Google Scholar]
- 8.Morin B, Rahmeh AA, Whelan SP. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 2012;31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noton SL, Deflubé LR, Tremaglio CZ, Fearns R. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog. 2012;8:e1002980. doi: 10.1371/journal.ppat.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan PC, et al. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018;14:e1006889. doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: Theoretical assignment of functional domains. J Gen Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 12.Liang B, et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein T, et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol. 2018;3:1084–1089. doi: 10.1038/s41564-018-0227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoenen T, et al. Complete genome sequences of three ebola virus isolates from the 2014 outbreak in west Africa. Genome Announc. 2014;2:e01331-14. doi: 10.1128/genomeA.01331-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.England TE, Uhlenbeck OC. 3′-terminal labelling of RNA with T4 RNA ligase. Nature. 1978;275:560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- 16.Zhuang F, Fuchs RT, Sun Z, Zheng Y, Robb GB. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 2012;40:e54. doi: 10.1093/nar/gkr1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raabe CA, Tang TH, Brosius J, Rozhdestvensky TS. Biases in small RNA deep sequencing data. Nucleic Acids Res. 2014;42:1414–1426. doi: 10.1093/nar/gkt1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paweska JT, et al. Experimental inoculation of Egyptian fruit bats (Rousettus aegyptiacus) with Ebola virus. Viruses. 2016;8:E29. doi: 10.3390/v8020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones ME, et al. Experimental inoculation of Egyptian rousette bats (Rousettus aegyptiacus) with viruses of the ebolavirus and marburgvirus genera. Viruses. 2015;7:3420–3442. doi: 10.3390/v7072779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng M, et al. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. eLife. 2015;4:e11785. doi: 10.7554/eLife.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider U, Schwemmle M, Staeheli P. Genome trimming: A unique strategy for replication control employed by Borna disease virus. Proc Natl Acad Sci USA. 2005;102:3441–3446. doi: 10.1073/pnas.0405965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habjan M, et al. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vulliémoz D, Roux L. “Rule of six”: How does the Sendai virus RNA polymerase keep count? J Virol. 2001;75:4506–4518. doi: 10.1128/JVI.75.10.4506-4518.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vulliémoz D, Roux L. Given the opportunity, the Sendai virus RNA-dependent RNA polymerase could as well enter its template internally. J Virol. 2002;76:7987–7995. doi: 10.1128/JVI.76.16.7987-7995.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowton VM, Fearns R. Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3′ end of the nucleocapsid and can scan to access internal signals. J Virol. 2005;79:11311–11322. doi: 10.1128/JVI.79.17.11311-11322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn JH. 2008. Filoviruses: A Compendium of 40 Years of Epidemiological, Clinical, and Laboratory Studies, Archives of Virology, ed Calisher CH (Springer, New York), Vol 20.
- 28.Mühlberger E. Filovirus replication and transcription. Future Virol. 2007;2:205–215. doi: 10.2217/17460794.2.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cressey TN, Noton SL, Nagendra K, Braun MR, Fearns R. Mechanism for de novo initiation at two sites in the respiratory syncytial virus promoter. Nucleic Acids Res. 2018;46:6785–6796. doi: 10.1093/nar/gky480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mühlberger E, Weik M, Volchkov VE, Klenk H-D, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theriault S, Groseth A, Neumann G, Kawaoka Y, Feldmann H. Rescue of Ebola virus from cDNA using heterologous support proteins. Virus Res. 2004;106:43–50. doi: 10.1016/j.virusres.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Yang XL, et al. Characterization of a filovirus (Mengla virus) from Rousettus bats in China. Nat Microbiol. 2019;4:390–395. doi: 10.1038/s41564-018-0328-y. [DOI] [PubMed] [Google Scholar]
- 33.Manhart WA, et al. A chimeric Lloviu virus minigenome system reveals that the bat-derived filovirus replicates more similarly to ebolaviruses than marburgviruses. Cell Rep. 2018;24:2573–2580.e4. doi: 10.1016/j.celrep.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin A, Hoefs N, Tadewaldt J, Staeheli P, Schneider U. Genomic RNAs of Borna disease virus are elongated on internal template motifs after realignment of the 3′ termini. Proc Natl Acad Sci USA. 2011;108:7206–7211. doi: 10.1073/pnas.1016759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crary SM, Towner JS, Honig JE, Shoemaker TR, Nichol ST. Analysis of the role of predicted RNA secondary structures in Ebola virus replication. Virology. 2003;306:210–218. doi: 10.1016/s0042-6822(02)00014-4. [DOI] [PubMed] [Google Scholar]
- 36.Chen IH, et al. Characterization of the polyadenylation activity in a replicase complex from Bamboo mosaic virus-infected Nicotiana benthamiana plants. Virology. 2013;444:64–70. doi: 10.1016/j.virol.2013.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranjith-Kumar CT, et al. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: Implication for viral RNA synthesis. J Virol. 2001;75:8615–8623. doi: 10.1128/JVI.75.18.8615-8623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomar S, Hardy RW, Smith JL, Kuhn RJ. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol. 2006;80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu W, et al. Flock house virus RNA polymerase initiates RNA synthesis de novo and possesses a terminal nucleotidyl transferase activity. PLoS One. 2014;9:e86876. doi: 10.1371/journal.pone.0086876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen MW, et al. Chikungunya virus nsP4 RNA-dependent RNA polymerase core domain displays detergent-sensitive primer extension and terminal adenylyltransferase activities. Antiviral Res. 2017;143:38–47. doi: 10.1016/j.antiviral.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerlier D, Lyles DS. Interplay between innate immunity and negative-strand RNA viruses: Towards a rational model. Microbiol Mol Biol Rev. 2011;75:468–490. doi: 10.1128/MMBR.00007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimberlin CR, et al. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci USA. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcin D, et al. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marq JB, Kolakofsky D, Garcin D. Unpaired 5′ ppp-nucleotides, as found in arenavirus double-stranded RNA panhandles, are not recognized by RIG-I. J Biol Chem. 2010;285:18208–18216. doi: 10.1074/jbc.M109.089425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcin D, Kolakofsky D. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol. 1992;66:1370–1376. doi: 10.1128/jvi.66.3.1370-1376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jordan I, Horn D, Oehmke S, Leendertz FH, Sandig V. Cell lines from the Egyptian fruit bat are permissive for modified vaccinia Ankara. Virus Res. 2009;145:54–62. doi: 10.1016/j.virusres.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olejnik J, et al. Ebolaviruses associated with differential pathogenicity induce distinct host responses in human macrophages. J Virol. 2017;91:e00179-17. doi: 10.1128/JVI.00179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haddock E, Feldmann F, Feldmann H. Effective chemical inactivation of Ebola virus. Emerg Infect Dis. 2016;22:1292–1294. doi: 10.3201/eid2207.160233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fearns R, Peeples ME, Collins PL. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 52.Nelson EV, et al. An RNA polymerase II-driven Ebola virus minigenom system as an advanced tool for antiviral drug screening. Antiviral Res. 2017;146:21–27. doi: 10.1016/j.antiviral.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsen ME, Cressey TN, Mühlberger E, Connor JH. Differential mechanisms for the involvement of polyamines and hypusinated eIF5A in Ebola virus gene expression. J Virol. 2018;92:e01260-18. doi: 10.1128/JVI.01260-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trunschke M, et al. The L-VP35 and L-L interaction domains reside in the amino terminus of the Ebola virus L protein and are potential targets for antivirals. Virology. 2013;441:135–145. doi: 10.1016/j.virol.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fearns R, Collins PL. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J Virol. 1999;73:5852–5864. doi: 10.1128/jvi.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.