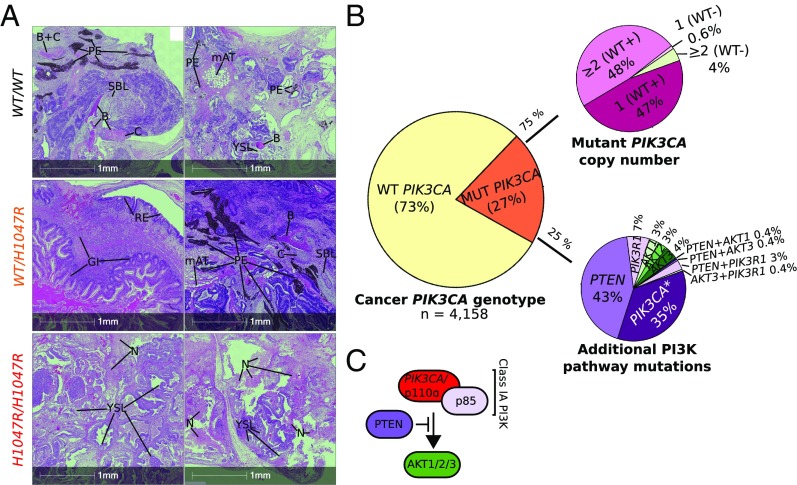
Fig. 7.
PIK3CAH1047R allele dose-dependent effects in tumor xenografts and genetic evidence for graded PI3K activation in cancers. (A) Hematoxylin and eosin (H&E)-stained sections of WT (WT/WT), PIK3CAWT/H1047R (WT/H1047R), and PIK3CAH1047R/H1047R (H1047R/H1047R) tumor xenografts derived from injection of hPSCs into immunodeficient mice. The micrographs are from two tumors per genotype and are representative of totals of five, three, and two tumors from WT, PIK3CAWT/H1047R, and PIK3CAH1047R/H1047R iPSCs, respectively. Yolk sac-like (YSL) and embryonal carcinoma-like (ECL) tissues, suggesting neoplastic transformation of cells within the original cultures, were more prevalent in PIK3CAH1047R/H1047R tumors, which also exhibited extensive necrosis (N); rare YSL foci were seen in two other tumors derived from the same WT clone. The only well-differentiated tissue observed in PIK3CAH1047R/H1047R tumors was a focus of immature bone (B) in one. WT and PIK3CAWT/H1047R tumors, in contrast, comprised variable admixtures of well-differentiated and organized tissue derivatives of all three germ layers. GI, gastrointestinal tissue; mAT, mouse adipose tissue (confirmed by independent mouse vs. human immunostaining with Cyclophilin A; SI Appendix, Fig. S8 A and B); PE, pigmented epithelium; RE, respiratory epithelium; SBLs, sebaceous-like tissue. See also SI Appendix, Fig. S8 and Table S1. (B) The Cancer Genome Atlas (TCGA) was used to extract genomic data from PIK3CA-associated cancers. These were analyzed in aggregate for the presence or absence of mutant PIK3CA alleles, followed by stratification of PIK3CA mutant-positive samples based on the presence of multiple mutant alleles, including cases where the WT PIK3CA allele is lost (WT−). Alternatively, PIK3CA mutant-positive samples were screened for multiple distinct PIK3CA mutations (*) or for the presence of additional mutations in proximal PI3K pathway components. (C) Schematic of proximal class IA PI3K signaling of relevance to the analysis in B.