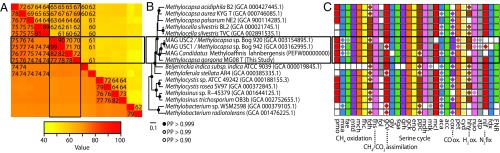
Fig. 2.
Average nucleotide and amino acid identities, phylogenetic relationship, and central metabolism comparison between M. gorgona MG08 and its genome sequenced relatives. (A) Symmetrical matrix of pairwise gANI and AAIs between all strains and MAGs and ordered as in B. ANI is presented in the Lower Left triangle and values ≥74 are provided. AAI is presented in the Upper Right triangle and values ≥60 are provided. M. gorgona MG08 and Ca. M. lahnbergensis (AAI, 71.3; ANI, 78.1), MAG USC1 (AAI, 70.1; ANI, 78.0), MAG USC2 (AAI, 70.0; ANI, 77.3), M. aurea KYG T (AAI, 69.3; ANI, 72.5), M. acidiphila B2 (AAI, 67.4; ANI, 74.75), M. palsarum NE2 (AAI, 66.1; ANI, 67.9) and M. silvestris BL2 (AAI, 62.5; ANI, 69.9) are below the species threshold of 96.6 ANI (3) and 95 AAI (4). (B) The phylogenomic tree was calculated with 10 independent chains of 11,000 generations under the LG model with four rate categories, using an input alignment of 34 concatenated marker genes (Materials and Methods). A total of 6,000 generations of each chain were discarded as burn-in, the remainder were subsampled every five trees (bpcomp -x 6000 5 11000) and pooled together for calculation of the reported 50% consensus tree and bipartition posterior probabilities (maxdiff = 0.814, meandiff = 0.010076). The model and number of rate categories was identified using ModelFinder (Materials and Methods). (C) Distribution of functional complexes presented in Fig. 4 and SI Appendix, Table S1 were determined using blast (5), OrthoFinder (6), and manual examination of trees. Presence of a complete complex is indicated by a solid square. Complexes that are incomplete are indicated with an embedded diamond. Abbreviations for functional complexes: aca, carbonic anhydrase; acc, acetyl-CoA carboxylase; atp, ATP synthase; cox, carbon monoxide dehydrogenase; cyd, terminal cytochrome oxidase; eno, enolase; fae, formaldehyde activating enzyme; fdh, formate dehydrogenase; fdx, ferredoxin, 2Fe-2S; fhc, formyltransferase/hydrolase complex; fhs, formate–tetrahydrofolateligase; FNR, ferredoxin-NADP+ oxidoreductase; fol, bifunctional 5,10-methylene-tetrahydrofolatedehydrogenase, and 5,10-methylene-tetrahydrofolatecyclohydrolase; gck, 2-glycerate kinase; gcv, glycine cleavage complex; gly, serine hydroxymethyltransferase; hhy, [NiFe] hydrogenase; hpr, hydroxypyruvate reductase; mch, methenyl tetrahydromethanopterin cyclohydrolase; mcl, malyl-CoA lyase; mdh, malate dehydrogenase; mtd, NAD(P)-dependent methylene tetrahydromethanopterin dehydrogenase; mtk, malate thiokinase; mxa, methanol dehydrogenase; nif, nitrogenase; nuo, NADH-quinone oxidoreductase; pet, ubiquinol-cytochrome c reductase; pmo, particulate methane monooxygenase; ppc, phosphoenolpyruvate carboxylase; and sga, serine–glyoxylate aminotransferase.