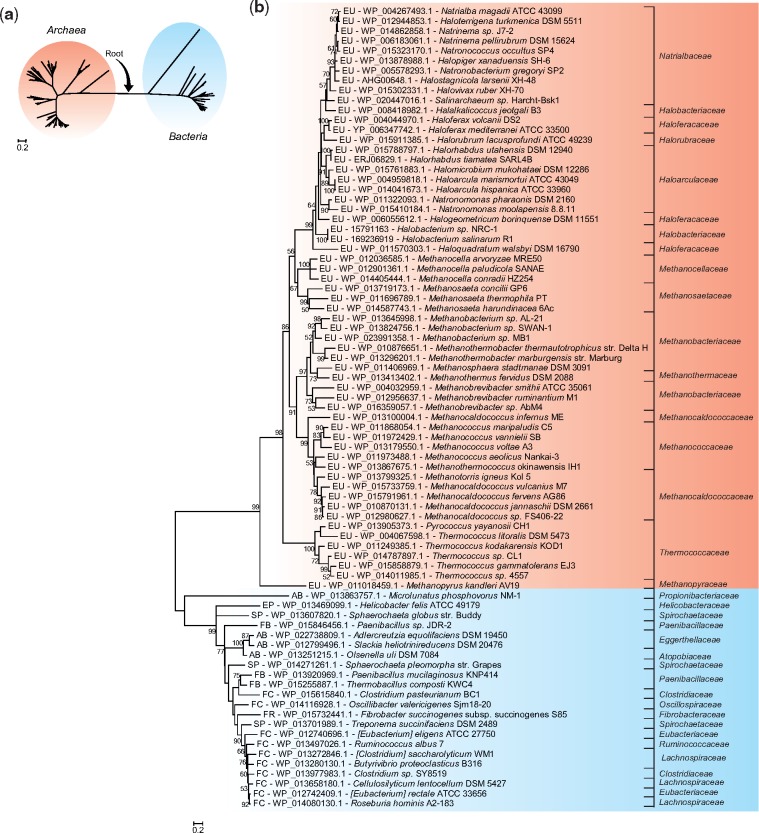
Fig. 3.
—Phylogenetic relationships among archaeal PurOs and their bacterial counterparts. (a) Maximum likelihood tree (ML) of PurOs showing the root placement at the branch that connects PurOs from Archaea and Bacteria. (b) Rooted ML tree showing PurOs from Archaea and their homologs in Bacteria in distinct groups. The multiple alignment utilized in these reconstructions contained 81 sequences of amino acids with 134 sites. The tree was constructed with the model LG+G + I and bootstrap was performed with 1,000 resamplings. The OTUs are identified by abbreviations that represent their phylum and class, accession number of the protein and species name. The abbreviations are: EU, phylum Euryarchaeota; FC, phylum Firmicutes class Clostridia; FB, phylum Firmicutes class Bacillales; AB, phylum Actinobacteria; SP, phylum Spirochaetes; FR, phylum Fibrobacteres; EP, phylum Proteobacteria classe Epsilonproteobacteria. The scale indicates the number of substitutions per site.