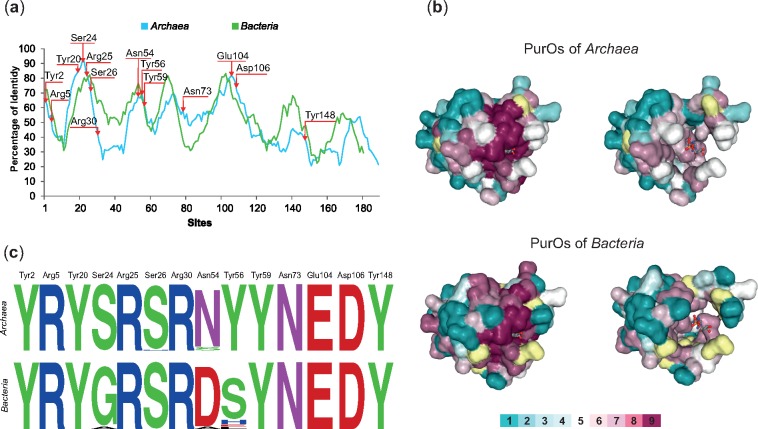
Fig. 4.
—Conservation of PurOs from Archaea and Bacteria. (a) Sliding window plot analysis of the multiple alignments of PurOs in Archaea and Bacteria show that the conservation in the primary structure is similar in these domains. The arrows indicate the positions of the active sites of PurO from Methanothermobacter thermoautotrophicus. (b) Amino acids residues of the 3D structure of PurO from M. thermoautotrophicus complexed with AICAR, its substrate (PDB ID code 2NTL) as visualized in ConSurf. The tertiary structure is presented using a surface-filled model and the AICAR in a ball-and-stick model. The amino acids residues are colored according to their ConSurf conservation scores calculated on the basis of multiple alignments of archaeal and bacterial PurOs. The color-coding bar varies from 1 to 9, where 9 is the most conserved and 1 most variable. Amino acid positions with low confidence are marked in yellow. Tertiary structures on the left show amino acids with scores 1 to 9 and the ones on the right show amino acids with scores 1 to 7. The figure reveals that most highly conserved amino acids (scores 8 and 9) compose or surround the active site of the enzyme. (c) Sequence logos of the positions in the multiple alignment that correspond to the active sites of PurO from M. thermoautotrophicus showing that most amino acids are conserved in Archaea and Bacteria.