Figure 3.
DPF2 Associates with Active Enhancers and the Oct4, Sox2, and Brg1 Network in ESCs
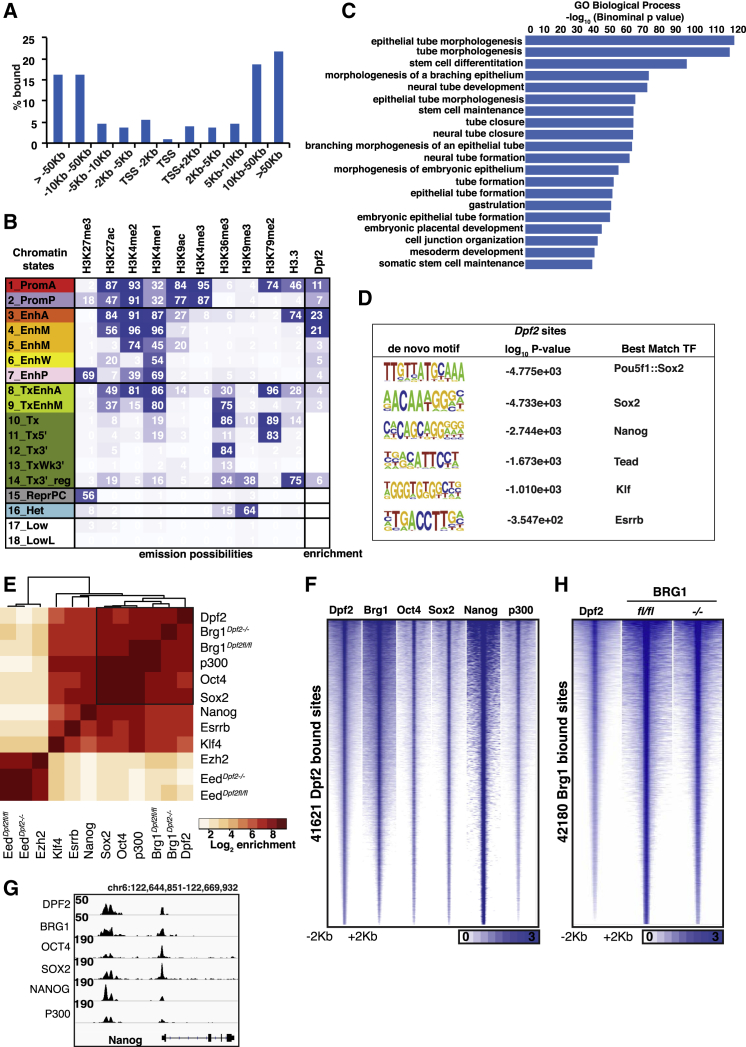
(A) Distribution of DPF2 target sites determined by ChIP-seq in ESCs in relation to their distance to TSSs.
(B) Chromatin state enrichment of DPF2 target sites in ESCs. ESC chromatin states were defined in Chronis et al. (2017) using ChromHMM. Rows represent chromatin states and their mnemonics. Columns give the frequency of the indicated histone marks and H3.3 for each chromatin state (ChromHMM emission probabilities), color-coded from blue (highest) to white (lowest). Enrichment of DPF2 in each chromatin state is shown in the last column.
(C) Significant GO terms for genes with DPF2 target sites within 10 kb of their TSS.
(D) Motifs identified at DPF2-bound sites by de novo search and the best matching TFs.
(E) Hierarchical clustering of pairwise enrichments of DPF2, BRG1, OCT4, SOX2, NANOG, ESRRB, KLF4, EED, and EZH2 binding sites in ESCs. BRG1 and EED binding sites were identified from Dpf2fl/fl and Dpf2−/− ESC lines, and all other binding events were obtained from WT ESCs. The black box indicates highest correlation enrichment for pairwise binding.
(F) Heatmaps of normalized ChIP-seq signal for Dpf2, Brg1, Oct4, Sox2, Nanog, and p300 at all sites bound by DPF2 in ESCs.
(G) Genome browser view of DPF2, BRG1, OCT4, SOX2, NANOG, and P300 binding at the Nanog locus in ESCs.
(H) Heatmaps of normalized ChIP-seq signal for Dpf2 and Brg1 at BRG1 occupied sites in Dpf2fl/fl or Dpf2−/− ESC lines.