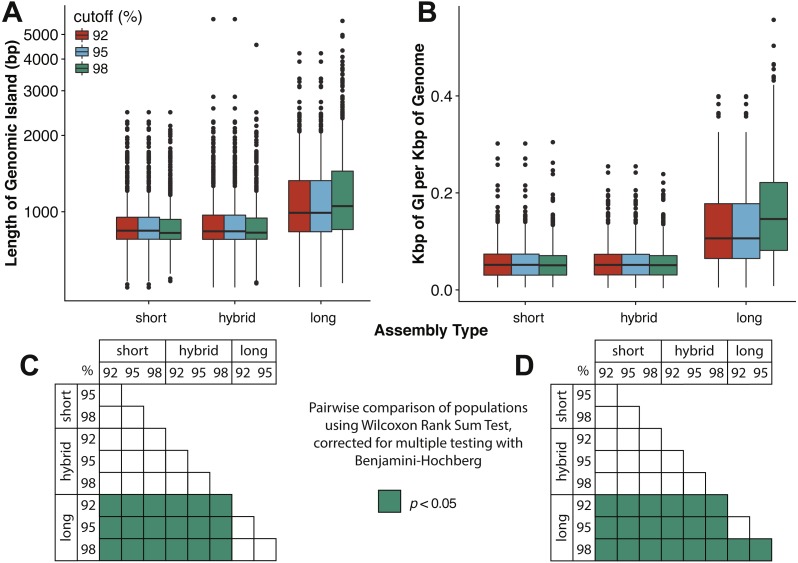
Figure 4. Long-read assemblies capture longer genomic islands than short-read methods.
Comparison of the (A) length of genomic islands (GI) and (B) normalised length of GI per kb of genome per contig captured on long read assemblies of VirION reads compared to short- read only and hybrid assemblies of viral contigs from the Western English Channel. Genomic islands were identified by mapping reads back against contig across a range of nucleotide percentage identities (92, 95, 98%) to account for residual error remaining in polished long- read assemblies. (C) and (D) represent pairwise significance calculated using a Wilcoxon Rank Sum Test, with p-values adjusted (Benjamini-Hochberg) for multiple testing, for (A) and (B), respectively. Effect sizes and 95% confidence intervals can be found in Fig. S4.