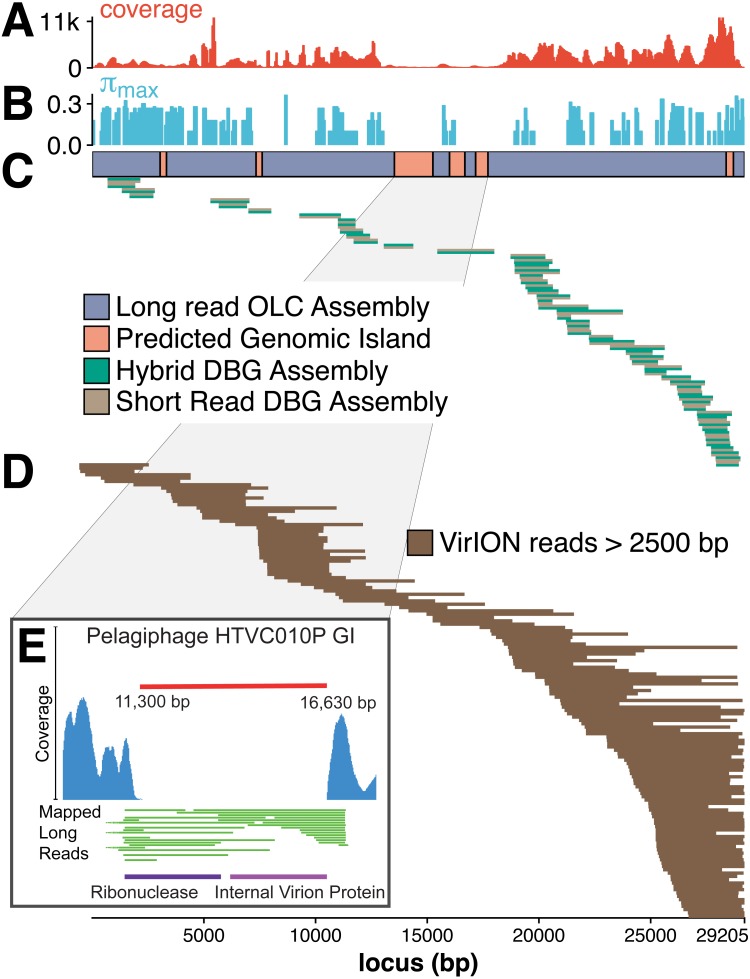
Figure 5. Long-read sequencing resolves microdiversity and assembly issues across genomic islands in ecologically important viral taxa.
De Bruijn Graph (DBG) assembly of short reads, even with VirION reads for scaffolding failed to assemble the genome of tig404, a virus closely related to the globally abundant pelagiphage HTVC010P. Only long-read assembly of VirION reads, followed by error correction with short read data was able to capture the complete genome on a single 29.2 kbp contig. A 200 bp sliding window analysis was used to calculate median coverage (A) of the assembly and (B) maximum nucleotide diversity (π), revealing six genomic islands (GIs) (C) and high levels of nucleotide diversity. The impact of this on short-read (light brown) only and hybrid assembly (green) can be seen in (C), where the assemblies aligned to the long-read assembly are highly fragmented. Conversely, long VirION reads (dark brown) were capable of spanning these regions across the whole genome and thus enabling assembly (D). One genomic island on tig404 was conserved with that of HTVC010P (E). Thus, we were able to identify the genomic content of this island at the population level by mapping VirION reads to HTVC010P and identifying those that spanned the genomic island. Encoded function was then predicted using tBLASTx to overcome high sequencing error in uncorrected VirION reads.