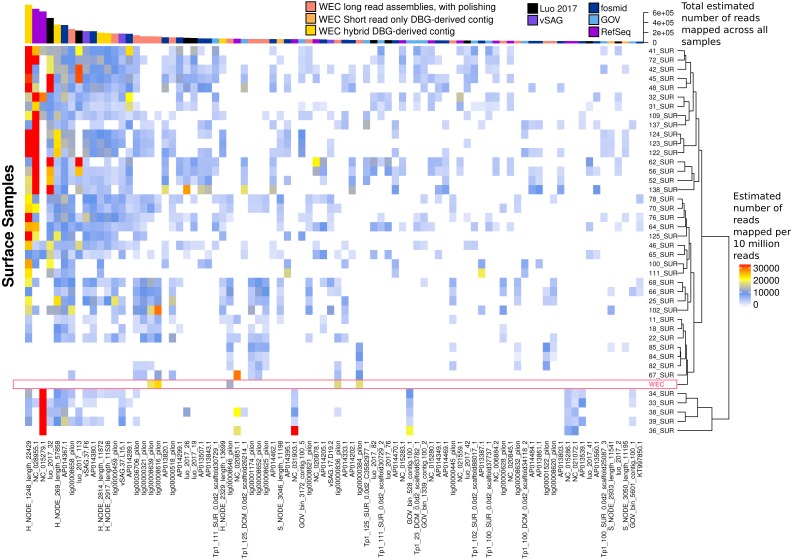
Figure 7. Ubiquity of VirION-derived Western English Channel viruses in Global Ocean surface waters.
Heatmap shows the top 50 most abundant and ubiquitous (appear in >10% of samples) viral contigs in the surface samples of the Global Ocean Virome (Roux et al., 2016a). Competitive recruitment of 10 million subsampled short reads was performed using FastViromeExplorer (Tithi et al., 2018) against a contig database comprising: (1) viral population contigs from this study; (2) viral genomes derived from other key viral metagenomic studies (Table S2); (3) Viruses from the NCBI RefSeq database. Estimated abundances are calculated from the total number of reads mapped to a contig, with reads mapping to multiple contigs apportioned to a single contig via an expectation-maximum algorithm (Tithi et al., 2018). Matrix columns are ordered (left to right) by total number of mapped reads across all samples. The most abundant contig was H_NODE_1248, which is related at the genus level to the ubiquitous pelagiphage vSAG-37-F6. The Western English Channel sample is highlighted in a pink box, showing globally ubiquitous and abundant viruses from oceanic provinces were not particularly abundant in this coastal sample.