Figure 3. The diverse pathways to the analogs of GA and GX2.
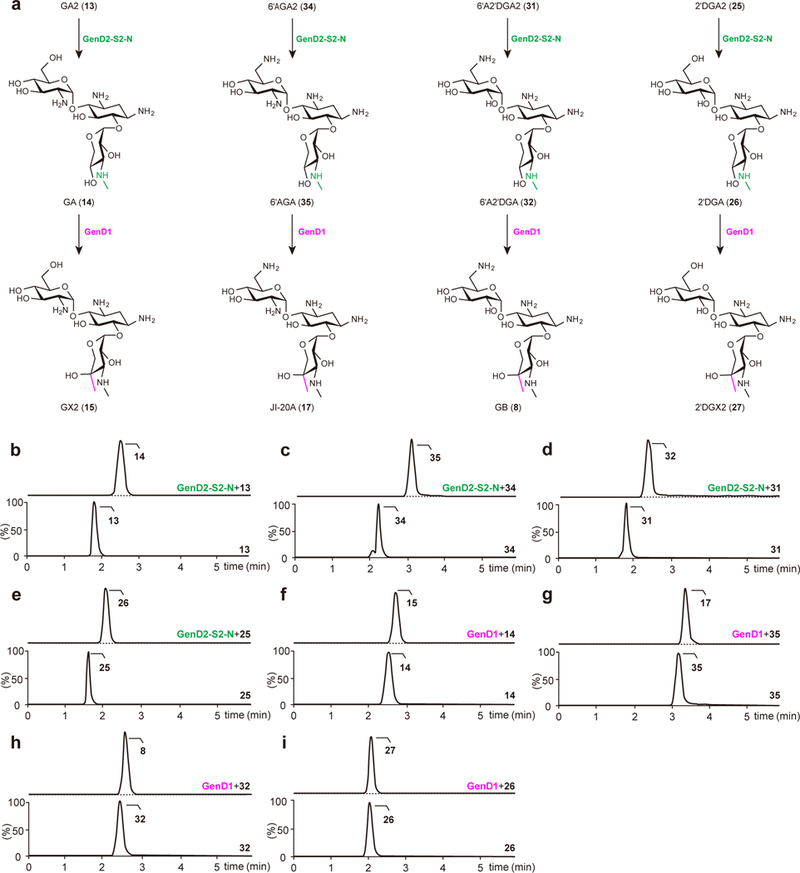
(a) C3″-methylamination steps to GA, 6′AGA, 6′A2′DGA, and 2′DGA catalyzed by GenD2-GenS2-GenN (indicated by green), and C4″ methylation steps to GX2, JI-20A, GB, and 2′DGX2 catalyzed by GenD1 (indicated by pink). (b–e) Chromatograms of GenD2-GenS2-GenN-catalyzed production of GA (selected for m/z = 469.2504) from GA2 (selected for m/z = 456.2188) (b), 6′AGA (selected for m/z = 468.2664) from 6′AGA2 (selected for m/z = 455.2348) (c), 6′A2′DGA (selected for m/z = 469.2504) from 6′A2′DGA2 (selected for m/z = 456.2188) (d), and 2′DGA (selected for m/z = 470.2344) from 2′DGA2 (selected for m/z = 457.2028) (e). (f–i) Chromatograms of GenD1-catalyzed production of GX2 (selected for m/z = 483.2661) from GA (f), JI-20A (selected for m/z = 482.2821) from 6′AGA (g), GB (selected for m/z = 483.2661) from 6′A2′DGA (h), and 2′DGX2 (selected for m/z = 484.2501) from 2′DGA (i). Lower chromatograms show the reactions without enzymes as controls. The dotted line in the upper chromatograms indicates the remaining substrate. Chromatograms show representative results of n>5 independent reactions.
