Abstract
Purpose
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been identified as a prognostic marker for the metastasis of early-stage non-small cell lung cancer (NSCLCs). We studied MALAT1 expression in breast cancer in relation to disease features and patient survival.
Methods
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was used to measure MALAT1 expression in tumor samples of 509 breast cancer patients. Hazards ratios (HRs) and 95% confidence intervals (CIs) were calculated to assess the association between MALAT1 expression and breast cancer survival using the Cox proportional hazards regression model, and the analysis was adjusted for age at surgery, tumor grade, disease stage, and hormone receptor status. Meta-analysis of multiple microarray datasets from online databases and our own study was performed to evaluate the association of MALAT1 with breast cancer survival.
Results
Patients with low grade or ER-positive tumors had higher expression of MALAT1 compared to those with high grade (p=0.013) or ER negative (p=0.0002) tumors. Patients with PR-positive tumors also had higher MALAT1 expression than those with PR-negative tumors (p<0.0001). In patients with positive hormone receptors or low tumor grade, tumors with high MALAT1 expression were more likely to recur. Survival analysis showed that patients with high expression of MALAT1 had a 2-fold increase in risk of relapse (p=0.0083) compared to those with low expression. This association remained significant after adjustment for age at surgery, disease stage, tumor grade and hormone receptor status. Meta-analysis showed that high MALAT1 expression was associated with poor relapse-free survival in patients with hormone receptor positive tumors (HR=1.44, 95%CI=1.08-1.92).
Conclusions
High expression of lncRNA MALAT1 is associated with breast cancer relapse and may play a role in tumor progression.
Keywords: MALAT1, Breast Cancer, Estrogen Receptor, Survival, Meta-analysis
Introduction
Breast cancer is the most common female malignancy in the US, accounting for nearly 30% of all new cancer diagnoses in women [1, 2]. Despite extensive research, the tumorigenic process of the breast remains elusive, and effective therapies available for patients with metastatic diseases are limited. More studies are needed to identify and characterize new genes and their products which promote tumor progression and metastasis. This knowledge will not only improve our understanding of the disease, but also help to develop new biomarkers or targets for disease prognosis and treatment. Breast cancer research in the past has focused largely on proteins and protein-coding genes, with limited attention to non-coding genes and their transcripts. Protein-coding genes account only for 2% of the human genome. If we consider that cancer is a genetic disease, it is conceivable that we are overlooking a large part of the genome which may be involved in the disease.
More than 75% of the human genome are transcribed, but a majority of the transcripts do not encode proteins, known as non-coding RNAs (ncRNAs) [3]. Studies have shown that dysregulated ncRNAs, including long non-coding RNAs (lncRNAs), play a crucial role in tumorigenesis and in determination of malignant phenotypes [4]. LncRNAs are defined as non-coding RNAs with more than 200 nucleotides in length and no potential for protein translation. These transcripts are found to regulate important biologic processes, such as development, differentiation and transformation [5–9], as well as to interact with other functional molecules, including DNA, proteins and microRNAs [10]. Evidence also suggests that many lncRNAs have tissue-specific expression and cell-specific function [7]. Subcellular localization also determines the function of lncRNA, as many lncRNAs are localized preferentially in the nucleus, regulating gene expression, like X inactive specific transcript (XIST), BMP2-OP1-responsive gene (BORG), and nuclear enriched abundant transcript 1 (NEAT1). LncRNAs are found frequently to be dysregulated in human cancer, and the dysregulation is associated with disease recurrence, metastasis and prognosis [11–13].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as noncoding nuclear-enriched abundant transcript 2 (NEAT2), is a lncRNA located on chr11q13 with more than 8.0 kb in length. MALAT1 was initially found to be a prognostic marker for the metastasis of early-stage non-small cell lung cancer (NSCLCs) [11]. MALAT1 is localized to nuclear speckles, and regulates messenger RNA splicing by interaction with SR proteins and splicing factors [14–16]. MALAT1 expression is upregulated in several solid tumors, including the breast, pancreas, lung, colon, prostate and liver [11], and is suspected to be involved in tumorigenesis or disease progression [17–20]. In the present study, we analyzed MALAT1 expression in breast cancer and investigated its associations with clinical and pathological features and patient survival.
Material and methods
Study patients
Patients diagnosed with primary breast cancer were recruited from two hospitals affiliated with the University of Torino, the University Hospital between January 1998 and July 1999 and Mauriziano Hospital between October 1996 and August 2012. Tumor samples were collected from 509 consented patients during surgical resection of their primary cancer. Most of the patients were followed after surgery for a median of 82.8 months, ranging from 1.6 to 196.4 months. Of these patients, 433 (85%) had information on relapse-free and overall survival. Patient clinical and pathological data were extracted from their medical records. The study was approved by the Ethic Review Committee at each hospital.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from fresh frozen breast tumor samples (~30 mg) using the Allprep DNA/RNA Kit (Qiagen). About 1 μg total RNA was reverse-transcribed into cDNA using the Reverse Transcription Kit from LifeTech. The cDNA was amplified for a specific region of MALAT1 after mixing with the SYBR Green Master Mix (LifeTech) and a pair of primers specific for MALAT1. GAPDH was used as reference. The PCR primers were designed using the sequence NR_002819.2 and synthesized by Integrated DNA Technologies. The primer sequences are: MALAT1 forward 5’-GTTCTGATCCCGCTGCTATT-3’ and reverse 5’-TCCTCAACACTCAGCCTTTATC-3’ and GAPDH forward 5’-GTCAAGGCTGAGAACGGGAA-3’ and reverse 5’-AAATGAGCCCCAGCCTTCTC-3’. PCR was run in the Roche LightCycler 480 system as previously described [12, 13]. Levels of MALAT1 expression were calculated as an expression index (EI) using the formula 1,000 × 2(−ΔCt), where ΔCt = Ct (MALAT1) − Ct (GAPDH). Each sample was analyzed in triplicate. Tests with poor replicates (coefficient of variation >15%) were repeated.
Meta-analysis of MALAT1 data
Datasets containing MALAT1 expression in breast cancer were extracted from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds). All the data were generated from the Affymetrix Human Genome U133 plus 2.0 array and U133A array. Ten datasets were identified with at least 50 patients in each, including GSE1456, GSE4922, GSE6532, GSE19615, GSE20711, GSE20685, GSE16446, GSE31448, GSE3494 and GSE42568. Expression data on probe 224567_x_at (which targets MALAT1) were used for meta-analysis. Breast cancer provisional data in The Cancer Genome Atlas (TCGA) were retrieved using the web-based tool cBioPortal (http://www.cbioportal.org/index.do) [21, 22]. Using the study-specific tertile distribution as cutoff, MALAT1 expression data were grouped into 3 categories named as “MALAT1_high” (MALAT1 expression >high tertile cutoff), “MALAT1_mid” (low tertile cutoff <= MALAT1 expression <high tertile cutoff), and “MALAT1_low” (MALAT1 expression <=low tertile cutoff). For the Cox proportional hazards regression analysis, MALAT1_ high was compared with MALAT1_low across the studies. In meta-analysis, pooled risk ratio (hazards ratio) and 95% confidence interval were calculated using the random-effect model (the DerSimonian and Laird method) which considers both within- and between-study variation [16, 23]. Review Manager (RevMan 5.3, Cochrane Collaboration) was used for meta-analysis.
Statistical analysis
Differences of MALAT1 expression classified by its tertile distribution (high, mid, and low) in relation to patient’s clinical and pathological parameters, including age at surgery, disease stage, tumor grade, status of estrogen receptor (ER) and progesterone receptor (PR) were analyzed using the Chi-square test. For survival analysis, hazards ratios (HRs) and 95% confidence intervals (CIs) were calculated using the Cox proportional hazards regression model with and without adjustment for age at surgery, tumor grade, disease stage, ER, and PR status. Overall survival (OS) was defined as the time interval from the date of surgery to the date of death or last follow-up. Relapse-free survival (RFS) was the time interval from surgery to recurrence or last follow-up. Statistical Analysis System software (version 9.4, SAS Institute) was used for all the statistical analyses. P values <0.05 (two tailed) were considered significant.
Results
Table 1 shows MALAT1 expression in association with clinical and pathological characteristics of breast cancer patients in our study. MALAT1 expression was higher in low than in high grade tumors (p=0.013) and in ER positive than ER negative tumors (p=0.0002). Patients with PR-positive tumors also had higher MALAT1 expression than those with PR-negative tumors (p<0.0001). However, compared to those without relapse, patients with relapse had higher expression of MALAT1 even though they had low grade tumors or ER positive diseases (data not shown). No significant differences were found in MALAT1 expression by age at surgery or disease stage.
Table 1.
Associations of MALAT1 expression with clinical and pathological factors of breast cancer
| Variables | Total No. (N=509) |
MALAT1 expression |
P value | ||
|---|---|---|---|---|---|
| Low No. (%) | Mid No. (%) | High No. (%) | |||
| Age at Surgery (year) | |||||
| ≤57.80 | 253 (49.90) | 88 (34.78) | 86 (33.99) | 79 (31.23) | 0.57 |
| >57.80 | 254 (50.10) | 78 (30.71) | 88 (34.65) | 88 (34.65) | |
| Disease Stage | 0.30 | ||||
| Stage 1 | 168 (34.93) | 49 (29.17) | 53 (31.55) | 66 (39.29) | |
| Stage 2 | 241 (50.10) | 83 (34.44) | 78 (32.37) | 80 (33.20) | |
| Stage 3 & 4 | 72 (14.96) | 24 (33.33) | 32 (44.44) | 16 (22.22) | |
| Tumor Grade | 0.013 | ||||
| Grade 1 | 60 (12.12) | 13 (21.67) | 19 (31.67) | 28 (46.67) | |
| Grade 2 | 192 (38.79) | 50 (26.04) | 65 (33.85) | 77 (40.10) | |
| Grade 3 | 243 (49.09) | 101 (41.56) | 83 (34.16) | 59 (24.28) | |
| ER Status | 0.0002 | ||||
| Positive | 349 (70.08) | 97 (27.79) | 122 (34.96) | 130 (37.25) | |
| Negative | 149 (29.92) | 68 (45.64) | 48 (32.21) | 33 (22.15) | |
| PR Status | <0.0001 | ||||
| Positive | 295 (59.36) | 75 (25.42) | 111 (37.63) | 109 (36.95) | |
| Negative | 202 (40.64) | 89 (44.06) | 59 (29.21) | 54 (26.73) | |
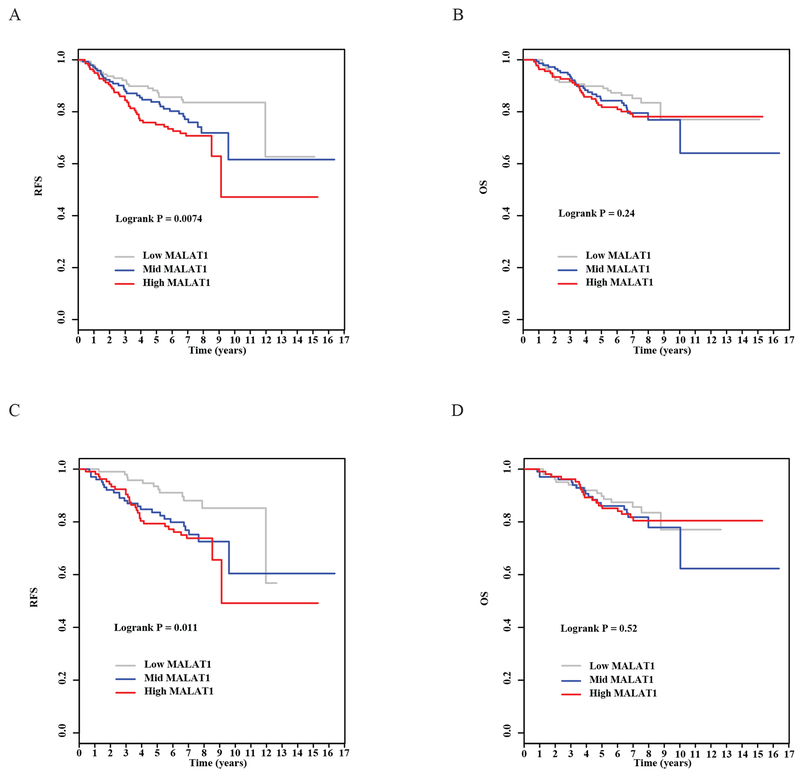
Survival analysis showed that patients with high expression of MALAT1 had a 2-fold increase in risk of relapse (HR=2.02, p=0.0083, Table 2) compared to those with low expression, and the survival curves were significantly different in patients with low, mid and high expression of MALAT1 (p=0.0074, Figure 1A). The association with relapse-free survival remained significant after patient age at surgery, tumor grade, disease stage, ER and PR status were adjusted in the analysis (HR=3.04, p=0.0001, Table 2). A linear correlation was also significant between MALAT1 expression and risk of relapse (HR=1.73, p<0.0001 after adjustment, Table 2). MALAT1 expression was not associated with overall survival in our univariate analysis (Table 2, Figure 1B), but the association became significant after we adjusted for the clinical and pathological variables. Patients with high expression of MALAT1 had significantly increased risk of death compared to those with low expression (HR=2.51, p=0.0050, Table 2), and the increase in risk was dose-dependent (p=0.0046, Table 2).
Table 2.
Associations of MALAT1 expression with breast cancer survival
| Variable | RFS |
P value | OS |
P value | RFS* |
P value | OS* |
P value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | HR | 95% CI | |||||
| MALAT1 | ||||||||||||
| Low | 1 | 1 | 1 | 1 | ||||||||
| Mid | 1.56 | 0.91-2.67 | 0.11 | 1.34 | 0.76-2.36 | 0.31 | 1.88 | 1.06-3.33 | 0.03 | 1.53 | 0.80-2.92 | 0.20 |
| High | 2.02 | 1.20-3.40 | 0.0083 | 1.41 | 0.80-2.48 | 0.24 | 3.04 | 1.73-5.37 | 0.0001 | 2.51 | 1.32-4.77 | 0.0050 |
| Continuous | 1.41 | 1.09-1.81 | 0.0080 | 1.18 | 0.89-1.56 | 0.24 | 1.73 | 1.31-2.28 | <0.0001 | 1.59 | 1.15-2.19 | 0.0046 |
Adjusted for age at surgery, tumor grade, disease stage, ER, and PR.
Figure 1. Associations of MALAT1 expression with patient survival.
A) Kaplan-Meier estimates for relapse-free survival by high, mid, and low MALAT1 expression. B) Kaplan-Meier estimates for overall survival by high, mid, and low MALAT1 expression. C) Kaplan-Meier estimates for relapse-free survival by high, mid, and low MALAT1 expression in hormone receptor positive patients. D) Kaplan-Meier estimates for overall survival by high, mid, and low MALAT1 expression in hormone receptor positive patients.
Since high MALAT1 expression was associated with low tumor grade and hormone receptor-positive tumors, we performed additional survival analysis in subgroups of patients who were stratified by tumor grade (Grade 1 or 2 versus 3) or hormone receptor status (ER or PR positive versus ER and PR negative). Results of the stratified analyses are shown in Table 3. The association between high MALAT1 expression and poor relapse-free survival was observed consistently in two subgroups of patients classified by tumor grades, and the strength of the association appeared to be stronger in grade 1 and 2 tumors compared to grade 3 tumors. For hormone receptor status, the association was seen only in hormone receptor positive tumors; no significant association was found in hormone receptor negative tumors. For patients with low grade and hormone receptor positive tumors, MALAT1 had a significant influence on relapse-free survival; patients with high MALAT1 had higher risk for relapse than those with low MALAT1.
Table 3.
Associations of MATLA1 expression with breast cancer survival stratified by tumor grade and/or hormone receptor status
| MALAT1 | RFS |
P value | OS |
P value | RFS* |
P value | OS* |
P value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | HR | 95% CI | |||||
| Grade 1or 2 | ||||||||||||
| Low | 1 | 1 | 1 | 1 | ||||||||
| Mid | 4.17 | 1.53-11.35 | 0.0052 | 2.36 | 0.95-5.87 | 0.064 | 3.88 | 1.39-10.88 | 0.0099 | 3.46 | 1.20-0.02 | 0.022 |
| High | 3.45 | 1.26-9.45 | 0.016 | 1.56 | 0.61-4.03 | 0.36 | 2.64 | 1.26-9.76 | 0.016 | 2.41 | 0.83-7.03 | 0.11 |
| Continuous | 1.58 | 1.06-2.37 | 0.024 | 1.19 | 0.79-1.81 | 0.41 | 1.63 | 1.07-2.47 | 0.023 | 1.41 | 0.89-2.21 | 0.14 |
| Grade 3 | ||||||||||||
| Low | 1 | 1 | 1 | 1 | ||||||||
| Mid | 1.32 | 0.63-2.74 | 0.46 | 1.04 | 0.47-2.31 | 0.93 | 1.33 | 0.61-2.88 | 0.47 | 0.97 | 0.40-2.31 | 0.94 |
| High | 2.15 | 1.11-4.16 | 0.023 | 1.47 | 0.71-3.05 | 0.30 | 2.57 | 126-5.23 | 0.0094 | 2.02 | 0.91-4.49 | 0.085 |
| Continuous | 1.49 | 1.07-2.07 | 0.019 | 1.22 | 0.84-1.77 | 0.29 | 1.64 | 1.15-2.34 | 0.0067 | 1.46 | 0.96-2.21 | 0.075 |
| ER+ or PR+ | ||||||||||||
| Low | 1 | 1 | 1 | 1 | ||||||||
| Mid | 2.18 | 1.09-4.36 | 0.027 | 1.30 | 0.65-2.57 | 0.46 | 2.32 | 1.13-4.75 | 0.022 | 2.74 | 1.04-7.21 | 0.041 |
| High | 2.43 | 1.23-4.78 | 0.011 | 1.26 | 0.64-2.48 | 0.51 | 2.82 | 1.39-5.73 | 0.0042 | 2.46 | 0.97-6.28 | 0.059 |
| Continuous | 1.49 | 1.09-2.02 | 0.012 | 1.12 | 0.80-1.55 | 0.52 | 1.61 | 1.16-2.22 | 0.0042 | 1.45 | 0.96-2.18 | 0.075 |
| ER− and PR− | ||||||||||||
| Low | 1 | 1 | 1 | 1 | ||||||||
| Mid | 1.77 | 0.69-4.57 | 0.24 | 1.45 | 0.50-4.17 | 0.22 | 1.59 | 0.61-4.19 | 0.35 | 1.06 | 0.34-3.27 | 0.92 |
| High | 1.86 | 0.72-4.79 | 0.20 | 1.70 | 0.60-4.77 | 0.51 | 1.81 | 0.64-5.12 | 0.26 | 1.30 | 0.39-4.29 | 0.67 |
| Continuous | 1.34 | 0.85-2.09 | 0.21 | 1.29 | 0.78-2.14 | 0.32 | 1.34 | 0.81-2.22 | 0.26 | 1.14 | 0.63-2.09 | 0.66 |
| Grade 1or 2 and ER+ or PR+ | ||||||||||||
| Low | 1 | 1 | 1 | 1 | ||||||||
| Mid | 3.55 | 1.28-9.81 | 0.015 | 2.30 | 0.86-6.15 | 0.096 | 3.50 | 1.21-10.16 | 0.021 | 4.09 | 1.24-13.44 | 0.020 |
| High | 2.92 | 1.05-8.15 | 0.041 | 1.78 | 0.66-4.82 | 0.26 | 3.13 | 1.08-9.04 | 0.035 | 3.20 | 0.99-10.36 | 0.053 |
| Continuous | 1.50 | 0.99-2.29 | 0.058 | 1.26 | 0.81-1.97 | 0.30 | 1.58 | 1.01-2.48 | 0.046 | 1.56 | 0.95-2.57 | 0.078 |
Adjusted for age at surgery and disease stage, as well as tumor grade or ER/PR status when they were not used for stratification.
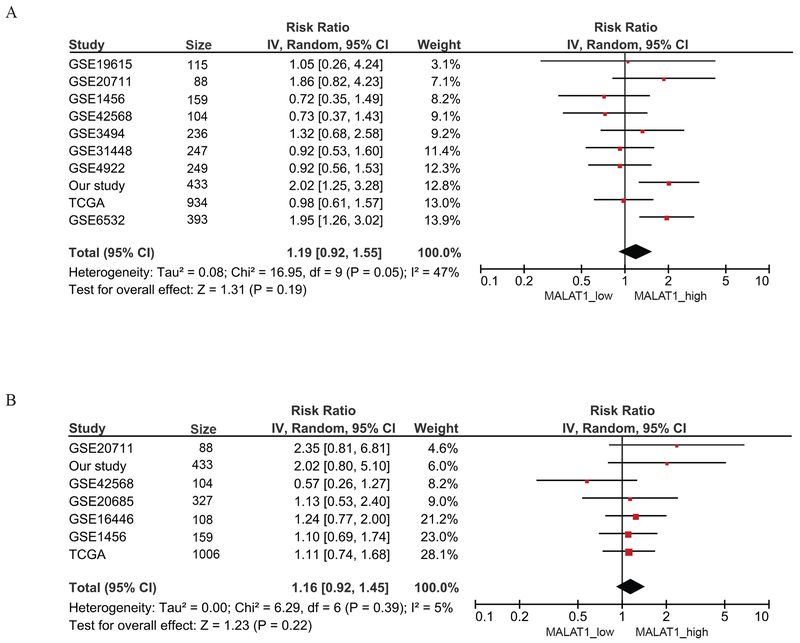
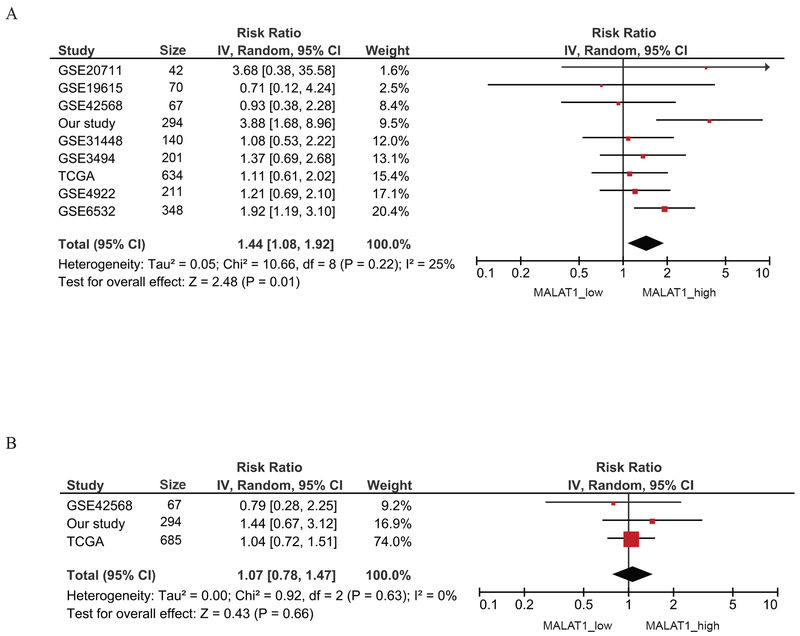
Meta-analysis of the association between MALAT1 expression and breast cancer survival was performed on 8 GEO datasets plus the TCGA breast cancer provisional data and our study. A total of 10 datasets with 2,958 patients had information on relapse-free survival, and 7 studies with 2,225 patients had overall survival data. The summarized results showed slightly increased risks of relapse and death for patients with high expression of MALAT1, but none of the associations were statistically significant (relapse-free survival HR=1.19, 95%CI: 0.92-1.55, Figure 2A; overall survival HR=1.16, 95%CI: 0.92-1.45, Figure 2B). However, when survival analysis was performed only in ER positive patients, MALAT1 expression was significantly associated with relapse-free survival. ER positive patients with high MALAT1 had a 44% higher risk (95% CI: 1.08-1.92) for relapse compared to ER positive patients with low expression (Figure 3A). The association for overall survival was not significant (Figure 3B).
Figure 2. Meta-analysis of the association between MALAT1 expression and breast cancer survival.
A) Meta-analysis of the association between MALAT1 expression and relapse-free survival. B) Meta-analysis of the association between MALAT1 expression and overall survival.
Figure 3. Meta-analysis of the association between MALAT1 expression and breast cancer survival in ER-positive patients.
A) Meta-analysis of the association between MALAT1 expression and relapse-free survival in ER-positive patients. B) Meta-analysis of the association between MALAT1 expression and overall survival in ER-positive patients.
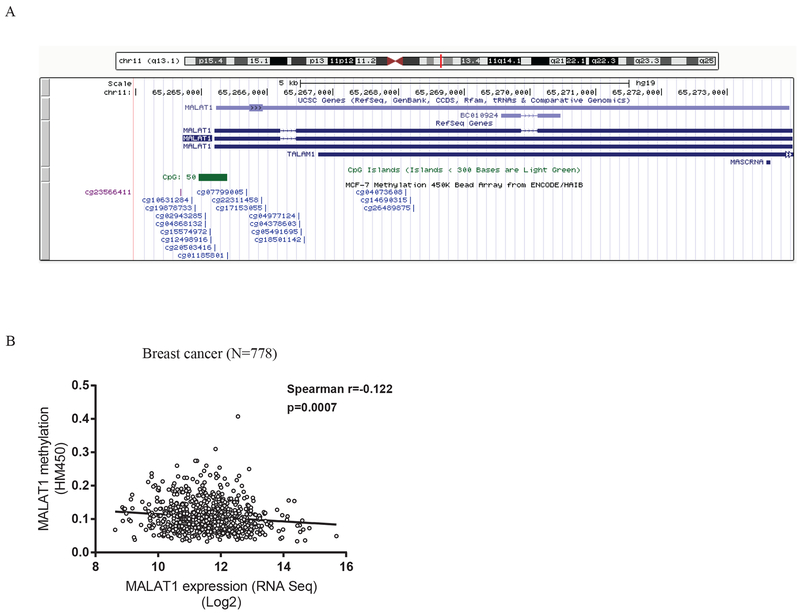
A CpG island was indicated in the genomic database annotated using the UCSC genome browser in an ER-positive cell line (MCF-7 cells) (Figure 4A). The relationship between MALAT1 expression and DNA methylation of the gene was evaluated after we merged the expression and methylation data available from TCGA where 778 breast cancer patients had gene expression data by RNA sequencing and DNA methylation data by the HumanMethylation450 chip. There were 10 methylation probes designed for the CpG sites in the promoter region of MALAT1, and 5 of them were in the CpG island. Overall, methylation levels in these CpG sites were very low, less than 0.1, and a weak inverse correlation was suggested between methylation and expression of MALAT1 (Spearman correlation coefficient=−0.122, p=0.00070, Figure 4B).
Figure 4. Methylation status and MALAT1 expression.
A) A screenshot from the UCSC Genome Browser shows the CpG island in the MALAT1 promoter and probes included in the Illumina HumanMethylation450 BeadChip for measuring methylation in the CpG sites. B) Scatter plot shows the correlation between MALAT1 expression and promoter methylation in breast cancer using the TCGA data.
Discussion
In the study, we found that MALAT1 levels in breast tumors were associated with breast cancer relapse and the association was independent of the established prognostic indicators, such as tumor grade, disease stage, and hormone receptor status. After adjustment for covariates, patients with high MALAT1 expression had a 3-fold increase in risk of relapse compared to those with low expression, and the increase in risk was significantly correlated with the level of expression. It was interesting to note that MALAT1 expression did not seem to be associated with overall survival in univariate analysis, but after adjusting for tumor grade, disease stage and receptor status, a significant association was detected. Patients with high MALAT1 had increased risk for death, which was consistent with the association for relapse. A significant association found after adjustment was quite unusual, suggesting that some covariates in the multivariate analysis may confound the relationship of MALAT1 and overall survival. Among the adjusted variables, we knew that tumor grade and hormone receptor status were associated with MALAT1 expression, and their associations with MALAT1 were in an opposing direction as to the association of MALAT1 with overall survival, i.e., high MALAT1 expression associated with poor survival, but meanwhile low grade or hormone receptor positive tumors having high expression of MALAT1. To address the confounding effect, we evaluated the survival associations in subgroups of patients stratified by tumor grade or hormone receptor status, and the subgroup analyses showed that the MALAT1’s associations with survival were more evident in patients with favorable prognostic indicators (low or mid tumor grades or ER or PR positive tumors) than in those with unfavorable indicators (high tumor grade or ER and PR negative tumors). The observation suggests that MALAT1 may have prognostic values only for certain patients. This limited value in prognosis may also explain why we did not find a consistent association between MALAT1 and survival in our meta-analysis when we did not stratify the patients by their tumor grade or hormone receptor status. To test this possibility, we performed meta-analysis only on ER-positive patients, and the analysis showed that MALAT1 expression was significantly associated with relapse-free survival, suggesting that MALAT1 may have prognostic values mainly in patients with hormone receptor positive tumors.
We also compared the median values of MALAT1 expression between patients with and without relapse in groups classified by tumor grade or ER status. In each group of comparisons, MALAT1 levels were higher in patients with relapse than in those without relapse, suggesting that MALAT1 may provide additional value of prognosis for patients who were initially considered to have good prognosis. Information on adjuvant chemo or hormonal therapy was available for 300 patients. We analyzed the treatment data with regard to the relationship between MALAT1 expression and treatment response. Our analysis showed that MALAT1 levels were not different between patients who responded and who did not respond, but for those who responded to adjuvant treatment low MALAT1 rendered better relapse-free survival compared to high MALAT1. These results were consistent with the earlier observations that high levels of MALAT1 in breast tumors were related to poor disease outcomes and that these relationships existed largely in patients who had a less aggressive disease or responded to adjuvant treatment.
Although MALAT1 is one of the lncRNAs discovered first in cancer, our understanding of the lncRNA’s functions is still limited. The MALAT1 gene is highly conserved across mammals, suggesting that it may have important biological implications [9]. Evidence suggests that MALAT1 may act like an oncogene in several malignancies, including lung cancer [17, 24], pancreatic cancer [19], liver cancer [20], bladder cancer [25], prostate cancer[26], colon cancer [27, 28], renal cell carcinoma [18], and oral squamous cell carcinomas [29, 30]. Lowering MALAT1 expression in hepatocellular carcinoma cells resulted in reduced cell proliferation and colony formation [20]. MALAT1 was also found to promote colon cancer cell proliferation, invasion and metastasis in vitro and in vivo by increasing AKAP-9 expression [28]. For breast cancer, suppressing MALAT1 expression led to increased differentiation of primary tumor cells and reduced metastasis [31]. Huang et al. reported that MALAT1 expression was elevated in breast tumors compared to adjacent normal tissues and high expression was associated with ER or PR positive tumors as well as poor disease-free survival, findings very similar to our observations in the current study [32].
Many lncRNAs dysregulated in human cancer were involved in the signal pathways of oncogenes or tumor suppressor genes [33]. For example, human maternally expressed gene 3 (MEG3) could inhibit cell proliferation through both p53-dependent and p53-independent pathways [34]. MALAT1 was found to be involved in the epithelial-mesenchymal transition (EMT)-mediated metastasis in oral squamous cell carcinoma by modulating the activation of β-catenin and NF-κB pathways [29]. MALAT1 could also regulate the Wnt-β-catenin pathway by enhancing nuclear β-catenin levels and promoting c-Myc expression [18, 31]. Furthermore, MALAT1 was reported to activate the mammalian target of rapamycin (mTOR) signaling pathway by regulating the alternative splicing of S6K1 [19]. MALAT1 regulated cell proliferation and metastasis in gallbladder carcinoma through activating the MAPK-ERK signaling pathway [35].
Using the TCGA data and Ingenuity Pathway Analysis (IPA), we performed pathway enrichment analysis based on the genes correlated with MALAT1 expression. The results of our analysis suggest that an active glycolytic pathway may be involved in the function of MALATI in breast cancer. Targeting tumor cell metabolisms and their key regulatory enzymes has emerged as an alternative strategy to complement the conventional genotoxic stress-based cancer therapy [36–38]. Studies have shown that cancer cells rely on glycolysis for energy production, whereas normal cells depend on the oxidative pathway [39, 40]. Highly proliferating tumor cells display an altered, high glycolytic metabolism called the Warburg effect [40, 41]. To date, a few studies have reported the role of lncRNAs in regulation of cancer metabolism. Prostate cancer gene expression marker 1 (PCGEM1) which is an androgen-induced prostate-specific lncRNA could increase cancer cell proliferation by enhancing aerobic glycolysis [42]. Li et al. reported that lncRNA urothelial cancer-associated 1 (UCA1) promoted glycolysis by upregulating hexokinase 2 (HK2) in bladder cancer cells [43]. Yang et al. found that hypoxia-induced LincRNA-p21 was critical for hypoxia-enhanced glycolysis [44]. Another study indicated that colorectal neoplasia differentially expressed (CRNDE) nuclear transcripts upregulated in colorectal cancer were involved in the regulation of cell metabolism [45]. Zhao et al. reported that LINC00092 promoted metastasis by altering glycolysis and sustaining the local supportive function of cancer-associated fibroblasts (CAFs) through binding to glycolytic enzyme-the fructose-2,6-biphosphatase PFKFB2 [46]. Knockdown lncRNA ceruloplasmin (NRCP) could significantly decrease glycolysis and increases mitochondrial respiration in ovarian cancer cells [47]. These studies suggest that lncRNAs may play an important role in cancer cell metabolism and targeting lncRNAs or their regulated metabolisms may offer new therapeutic applications.
DNA methylation is one of the epigenetic mechanisms that regulate gene expression in mammals [48]. Growing evidence demonstrates that DNA methylation in the promoter of a lncRNA gene can regulate its expression [49–52]. A CpG island was indicated in the MALAT1 promoter. To estimate if the CpG island plays a role in epigenetic regulation of MALAT1, we merged the DNA methylation and gene expression data from two breast cancer datasets in TCGA, and evaluated if MALAT1 expression was correlated with promoter methylation in breast cancer. Our analysis suggested a possible inverse correlation between MALAT1 methylation and expression, but the relationship was very weak, probably due to low methylation in the MALAT1 promoter in breast tumor tissues. Given the low level of methylation, we consider that DNA methylation may not play a role in regulation of MALAT1 expression in breast cancer.
In summary, we found that MALAT1 expression in breast cancer was paradoxically associated with tumor grade, ER/PR status and patient survival. While low grade or hormone receptor positive tumors were more likely to express MALAT1, high expression was associated with poor relapse-free survival. MALAT1 levels in breast tumors are indicative of breast cancer prognosis, but the prognostic value may be limited to patients with ER positive tumors.
Acknowledgment
University of Hawaii Cancer Center supported the study. Dr. Yuki Obata was supported by the Kinjo Gakuin University-Parent Teacher Association Research Grant.
Footnotes
Disclosure of potential conflicts of interest
None
Research involving Human Participants and/or Animals
The study involved human participants, but not animals
Informed consent
Study participants in our study provided informed consents
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer Statistics, 2017, CA Cancer J Clin, 67 (2017) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A, Breast cancer statistics, 2017, racial disparity in mortality by state, CA Cancer J Clin, 67 (2017) 439–448. [DOI] [PubMed] [Google Scholar]
- [3].Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR, Landscape of transcription in human cells, Nature, 489 (2012) 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schmitt AM, Chang HY, Long Noncoding RNAs in Cancer Pathways, Cancer Cell, 29 (2016) 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fatica A, Bozzoni I, Long non-coding RNAs: new players in cell differentiation and development, Nat Rev Genet, 15 (2014) 7–21. [DOI] [PubMed] [Google Scholar]
- [6].Wang KC, Chang HY, Molecular mechanisms of long noncoding RNAs, Mol Cell, 43 (2011) 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R, The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression, Genome Res, 22 (2012) 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL, Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses, Genes Dev, 25 (2011) 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES, Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals, Nature, 458 (2009) 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Morris KV, Mattick JS, The rise of regulatory RNA, Nat Rev Genet, 15 (2014) 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C, MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer, Oncogene, 22 (2003) 8031–8041. [DOI] [PubMed] [Google Scholar]
- [12].Shen Y, Katsaros D, Loo LW, Hernandez BY, Chong C, Canuto EM, Biglia N, Lu L, Risch H, Chu WM, Yu H, Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer, Oncotarget, 6 (2015) 8579–8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fu Y, Biglia N, Wang Z, Shen Y, Risch HA, Lu L, Canuto EM, Jia W, Katsaros D, Yu H, Long non-coding RNAs, ASAP1-IT1, FAM215A, and LINC00472, in epithelial ovarian cancer, Gynecol Oncol, 143 (2016) 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wilusz JE, Freier SM, Spector DL, 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA, Cell, 135 (2008) 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A, A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression, EMBO J, 29 (2010) 3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Larsson SC, Giovannucci E, Wolk A, Folate and risk of breast cancer: a meta-analysis, J Natl Cancer Inst, 99 (2007) 64–76. [DOI] [PubMed] [Google Scholar]
- [17].Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zornig M, MacLeod AR, Spector DL, Diederichs S, The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells, Cancer Res, 73 (2013) 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R, Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205, Cancer Res, 75 (2015) 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, Ji L, Kong R, Wang G, Jia YH, Bai XW, Sun B, Long Noncoding RNA MALAT1 Promotes Aggressive Pancreatic Cancer Proliferation and Metastasis via the Stimulation of Autophagy, Mol Cancer Ther, 15 (2016) 2232–2243. [DOI] [PubMed] [Google Scholar]
- [20].Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV, Karni R, Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation, Cancer Res, 77 (2017) 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal, Sci Signal, 6 (2013) pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Cancer Discov, 2 (2012) 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].DerSimonian R, Laird N, Meta-analysis in clinical trials, Control Clin Trials, 7 (1986) 177–188. [DOI] [PubMed] [Google Scholar]
- [24].Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC, Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression, Mol Cancer, 16 (2017) 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y, TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12, Clin Cancer Res, 20 (2014) 1531–1541. [DOI] [PubMed] [Google Scholar]
- [26].Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y, Zhang W, Jing T, Zhang C, Shen J, Xu C, Wang H, Wang H, Wang Y, Liu B, Li Y, Fang Z, Guo F, Qiao M, Wu C, Wei Q, Xu D, Shen D, Lu X, Gao X, Hou J, Sun Y, Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer, Oncotarget, 5 (2014) 11091–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG, MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9, Biochim Biophys Acta, 1852 (2015) 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hu ZY, Wang XY, Guo WB, Xie LY, Huang YQ, Liu YP, Xiao LW, Li SN, Zhu HF, Li ZG, Kan H, Long non-coding RNA MALAT1 increases AKAP-9 expression by promoting SRPK1-catalyzed SRSF1 phosphorylation in colorectal cancer cells, Oncotarget, 7 (2016) 11733–11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, Wu Y, Mei M, Zhang L, Wang X, Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma, Sci Rep, 5 (2015) 15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fang Z, Zhang S, Wang Y, Shen S, Wang F, Hao Y, Li Y, Zhang B, Zhou Y, Yang H, Long non-coding RNA MALAT-1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins, BMC Cancer, 16 (2016) 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, Spector DL, Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss, Genes Dev, 30 (2016) 34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huang NS, Chi YY, Xue JY, Liu MY, Huang S, Mo M, Zhou SL, Wu J, Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer, Oncotarget, 7 (2016) 37957–37965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tano K, Akimitsu N, Long non-coding RNAs in cancer progression, Front Genet, 3 (2012) 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A, Activation of p53 by MEG3 non-coding RNA, J Biol Chem, 282 (2007) 24731–24742. [DOI] [PubMed] [Google Scholar]
- [35].Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, Cao Y, Bao RF, Mu JS, Tan ZJ, Tao F, Liu YB, MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway, Cancer Biol Ther, 15 (2014) 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tennant DA, Duran RV, Gottlieb E, Targeting metabolic transformation for cancer therapy, Nat Rev Cancer, 10 (2010) 267–277. [DOI] [PubMed] [Google Scholar]
- [37].Vander Heiden MG, Targeting cancer metabolism: a therapeutic window opens, Nat Rev Drug Discov, 10 (2011) 671–684. [DOI] [PubMed] [Google Scholar]
- [38].Schulze A, Harris AL, How cancer metabolism is tuned for proliferation and vulnerable to disruption, Nature, 491 (2012) 364–373. [DOI] [PubMed] [Google Scholar]
- [39].Warburg O, On the origin of cancer cells, Science, 123 (1956) 309–314. [DOI] [PubMed] [Google Scholar]
- [40].Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation, Science, 324 (2009) 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Warburg O, On respiratory impairment in cancer cells, Science, 124 (1956) 269–270. [PubMed] [Google Scholar]
- [42].Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, Kung HJ, A long noncoding RNA connects c-Myc to tumor metabolism, Proc Natl Acad Sci U S A, 111 (2014) 18697–18702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li Z, Li X, Wu S, Xue M, Chen W, Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway, Cancer Sci, 105 (2014) 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yang F, Zhang H, Mei Y, Wu M, Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect, Mol Cell, 53 (2014) 88–100. [DOI] [PubMed] [Google Scholar]
- [45].Ellis BC, Graham LD, Molloy PL, CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism, Biochim Biophys Acta, 1843 (2014) 372–386. [DOI] [PubMed] [Google Scholar]
- [46].Zhao L, Ji G, Le X, Wang C, Xu L, Feng M, Zhang Y, Yang H, Xuan Y, Yang Y, Lei L, Yang Q, Lau WB, Lau B, Chen Y, Deng X, Yao S, Yi T, Zhao X, Wei Y, Zhou S, Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer, Cancer Res, 77 (2017) 1369–1382. [DOI] [PubMed] [Google Scholar]
- [47].Rupaimoole R, Lee J, Haemmerle M, Ling H, Previs RA, Pradeep S, Wu SY, Ivan C, Ferracin M, Dennison JB, Millward NMZ, Nagaraja AS, Gharpure KM, McGuire M, Sam N, Armaiz-Pena GN, Sadaoui NC, Rodriguez-Aguayo C, Calin GA, Drapkin RI, Kovacs J, Mills GB, Zhang W, Lopez-Berestein G, Bhattacharya PK, Sood AK, Long Noncoding RNA Ceruloplasmin Promotes Cancer Growth by Altering Glycolysis, Cell Rep, 13 (2015) 2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Klose RJ, Bird AP, Genomic DNA methylation: the mark and its mediators, Trends Biochem Sci, 31 (2006) 89–97. [DOI] [PubMed] [Google Scholar]
- [49].Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M, CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer, Oncogene, 29 (2010) 6390–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang L, Zhao Y, Bao X, Zhu X, Kwok YK, Sun K, Chen X, Huang Y, Jauch R, Esteban MA, Sun H, Wang H, LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration, Cell Res, 25 (2015) 335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, Ponting CP, The long non-coding RNA Dali is an epigenetic regulator of neural differentiation, Elife, 3 (2014) e04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D’Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG, DNMT1-interacting RNAs block gene-specific DNA methylation, Nature, 503 (2013) 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]