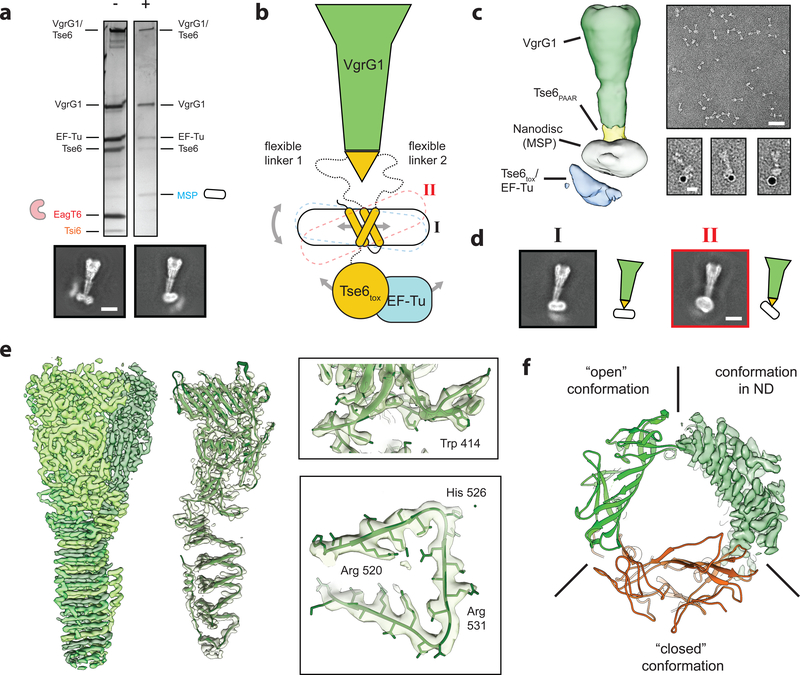
Figure 5. The toxin domain of Tse6 spontaneously crosses a lipid bilayer.
(a) Silver stained SDS-PAGE of the Tse6-loaded VgrG1 complex in its “pre-firing” conformation (−) and reconstituted in lipid nanodiscs (+). Upon reconstitution, EagT6 and Tsi6 dissociate from the complex and are exchanged by the nanodisc. Scale bar: 10 nm.
(b) Schematic representation of the VgrG1-Tse6-EF-Tu complex reconstituted in lipid nanodiscs. Flexibility caused by (1) lateral movement of the TMDs within the nanodisc, (2) tilting of the nanodisc as well as (3) movement of the Tse6tox-EF-Tu subcomplex impeded structural determination of the bottom part.
(c) Low-resolution cryo-EM reconstruction of VgrG1-Tse6-EF-Tu complex embedded in lipid nanodiscs (left) and representative negatively stained electron micrograph areas of the complex (right). Scale bar: 50 nm. Right panel shows three examples of VgrG1-Tse6-EF-Tu complexes in nanodiscs, labeled with 5 nm NTA-coated nanogold to label his-tagged Tse6. Scale bar: 10 nm.
(d) Two representative class averages of the VgrG1-Tse6-EF-Tu complex in side view (black I) and tilted view (red II), corresponding to the conformations shown in Fig. 5b. Scale bar: 10 nm.
(e) 3.2 Å cryo-EM reconstruction of VgrG1 obtained from the same dataset and applying C3 symmetry. Subunits of trimeric VgrG1 are colored in different green hues (left), fit of atomic model in single subunit (middle), as well as close-ups showing side chain densities (right) of the VgrG1 trimer.
(f) Comparison between atomic VgrG1 structures in ‘open’ and ‘closed’ conformations, showing that VgrG1 within the VgrG1-Tse6-EF-Tu complex in nanodiscs adopts an ‘open’ conformation.
See also Supplementary Figures 1–3 and 9 and Supplementary Tables 1–3 and Supplementary Video 2.