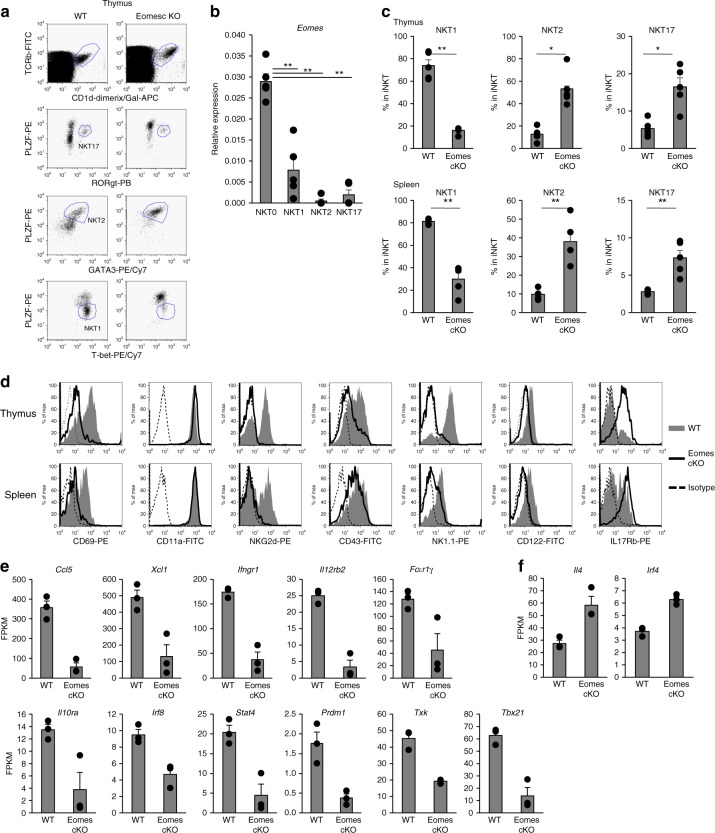
Fig. 2.
Aberrant expression of molecules affected by Eomes in iNKT terminal maturation and effector function. a Expression of PLZF, Rorγt, Gata3, and T-bet by thymic and splenic iNKT from WT and Eomes cKO mice detected by flow cytometry. b Quantitative PCR analysis of Eomes mRNA in thymic iNKT cells (TCRβ+CD1d-tetramer+) from WT mice: NKT0 (CD24+CD44−NK1.1−CD8−), NKT1 1(CD24−NK1.1+CD27+CCR6−), NKT2 (CD24−NK1.1-CD27+CD4+), NKT17 (CD24−CD27−CD4−CCR6+CD103+). (n = 5, mean ± SEM) **p < 0.01, two-tailed Student’s t-test. c The percentage of NKT1 (PLZFdimT-bet+), NKT2(PLZFhighGata3+), and NKT17(PLZF+Rorγt+) subsets in thymus and spleen of WT and Eomes cKO mice. (n = 5, mean ± SEM). *p < 0.05, **p < 0.01, Mann–Whitney. d Representative histograms showing expression of CD69, CD11a, NKG2d, CD43, NK1.1, CD122, and IL17Rb by thymic and splenic iNKT cells from WT and Eomes cKO mice (n = 5). Similar data were obtained from at least three independent experiments. e, f Thymic iNKT cells from WT and Eomes cKO mice were analyzed by RNA-Seq. Expression levels (expressed as FPKM) of selected NKT1 (e) or NKT2 (f) subset-specific genes of thymic iNKT cells were compared between WT and Eomes cKO mice (n = 3, mean ± SEM). (WT vs KO, *p < 0.05, **p < 0.01, ***< p < 0.001, two-tailed Student’s t-test)