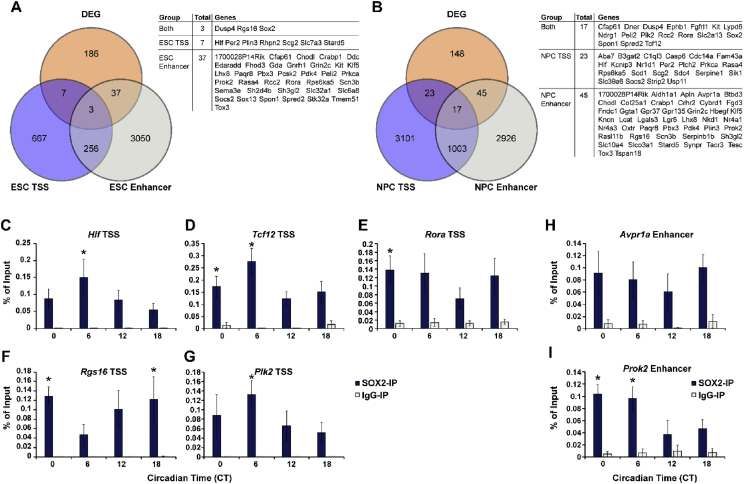
Fig. 5.
Identification of potential SOX2 target genes among the set of DEGs and their validation by ChIP-qPCR. (A–B) Venn diagrams illustrating the overlap between our DEG dataset and the SOX2 ChIP-seq datasets obtained from (A) ES cells and (B) NPCs, as reported in Lodato et al. (2013). The Lodato study distinguishes between SOX2 binding to TSS regions vs. binding to distal enhancer elements of the same gene. The tables list the genes that show SOX2 binding to both TSS and enhancer regions, TSS regions only, or enhancer regions only. (C–I) ChIP-qPCR analyses of the relative binding of SOX2 to the TSS regions of (C)Hlf, (D)Tcf12, (E)Rora, (F)Rgs16, and (G)Plk2, or to the distal enhancer regions of (H)Avpr1a and (I)Prok2 in SCN tissues harvested at 4 CTs. Values are represented as mean ± SEM. n = 3–6 per condition. *p<0.05 vs. IgG-IP.