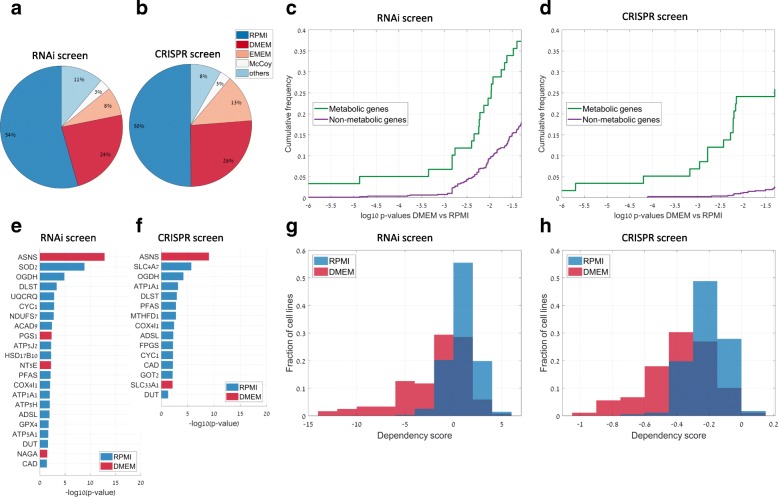
Fig. 1.
Tissue culture media significantly affect cancer cell dependence on metabolic enzymes. a, b The distribution of culture media types throughout cell lines used in large-scale RNAi genetic screen by Tsherniak et al. (a) and in a CRISPR-based screen by Meyers et al. (b). c, d Difference in RNAi (c)- and CRISPR (d)-based dependency scores of metabolic genes between cell lines grown in DMEM versus in RPMI (green) versus for other genes (purple), showing the cumulative distribution of FDR-corrected Wilcoxon p values. e, f Metabolic genes whose cell line-specific dependence is significantly correlated with the utilized tissue culture media in the RNAi (e) and CRISPR (f) screens. Blue and red represent genes that were found to be more essential in RPMI and DMEM, respectively. g, h The distribution of RNAi (g)- and CRISPR (h)-based dependency scores for ASNS (asparagine synthetase) throughout cell lines grown in DMEM versus in RPMI