Figure 2.
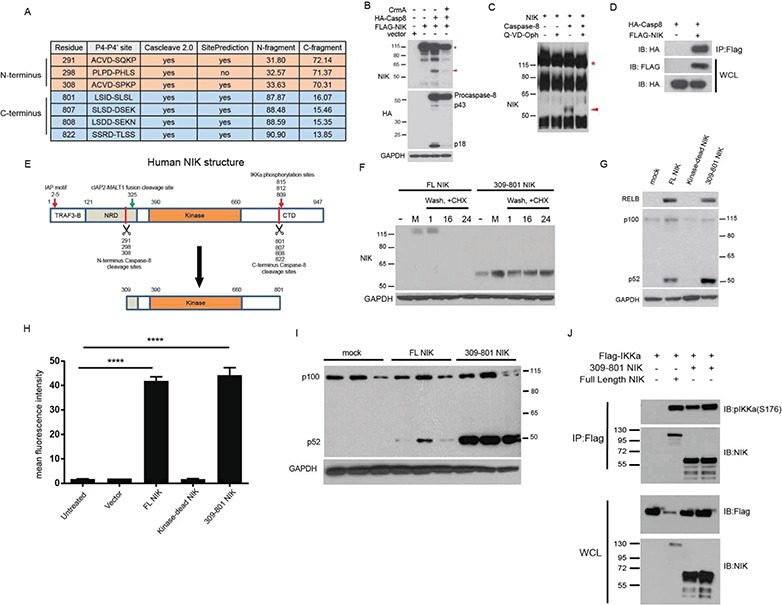
Caspase-8 cleaves NIK, resulting in a 55 kDa fragment of the kinase domain. (A) In silico prediction of human NIK cleavage by caspase-8 using Cascleave 2.0 and SitePrediction software programs. (B) IB of 293 T cells transfection experiment demonstrating that caspase-8 activity is sufficient for the cleavage of NIK to p55 NIK. (C) In vitro cleavage assay with purified NIK and recombinant caspase-8. (D) Co-immunoprecipitation experiment in 293 T cells with IP of FLAG-tagged NIK. (E) Structure of human NIK. TRAF3-B = TRAF3 binding domain, NRD = Negative Regulatory domain, Kinase= Kinase domain and CTD = C-terminal domain. (F) IB of 293 T cells transfected with the indicated plasmids. Cells were pulsed with proteasome inhibitor MG132, washed and incubated with the protein translation inhibitor CHX. Number labels indicate hour time course. (G) IB of 293 T cells transfected with the indicated plasmids. (H) Transfection experiment with Cignal-293 T cells in triplicate possessing an NF-κB responsive promoter upstream of a GFP reporter. ANOVA with Dunnett’s test for post hoc comparisons. ns = not significant, ****p < 0.0001; Error bars represent SEM. (I) Repeat of IB in (G) in triplicate showing increased processing of p100 to p52 in 309-801 NIK-transfected cells. (J) Co-immunoprecipitation experiment in 293 T cells with IP of FLAG-tagged IKKα. The red asterisk in (B) and (C) indicates full-length NIK, while the red arrowhead points to the 55 kDa fragment of the kinase domain. IB = Immunoblot, IP = Immunoprecipitation, WCL = whole cell lysate.
