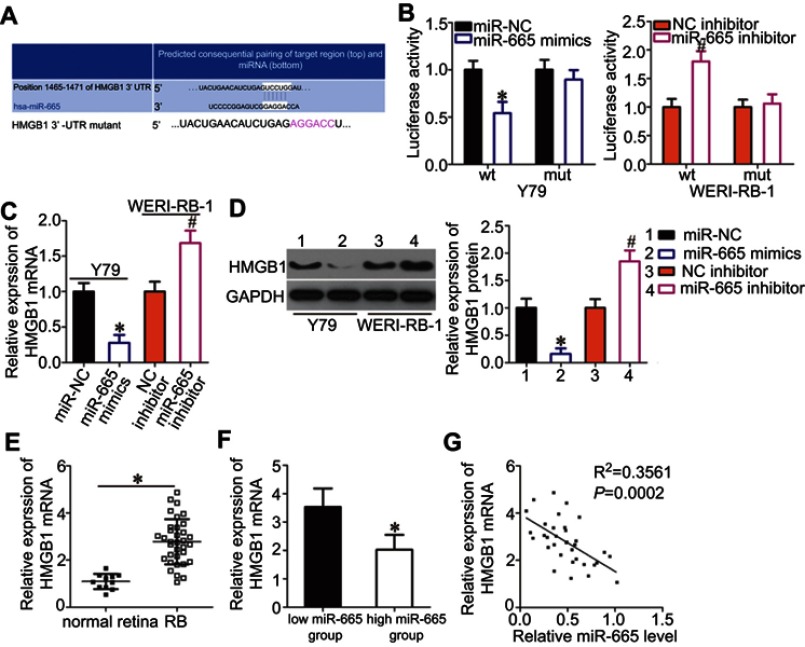
Figure 3.
Identification of HMGB1 as a direct target gene of miR-665 in RB cells. (A) Bioinformatics prediction revealed a highly conserved miR-665 binding site in the 3′-UTR of HMGB1. (B) Y79 cells were treated with either miR-665 mimics or miR-NC and either wild-type (wt) or mutant (mut) 3′-UTR reporter plasmid. miR-665 inhibitor or NC inhibitor along with the wt or mut 3′-UTR reporter plasmid were transfected into WERI-RB-1 cells. Luciferase activity was measured at 48 h post-transfection and normalized to that of the Renilla luciferase activity. *P<0.05 compared with miR-NC. #P<0.05 compared with NC inhibitor. (C, D) Expression levels of HMGB1 mRNA and protein in miR-665-overexpressing Y79 cells and miR-665-knocked down WERI-RB-1 cells were measured by using RT-qPCR and Western blot analysis, respectively. On day 15, a photo of colonies formed was taken. *P<0.05 compared with miR-NC. #P<0.05 compared with NC inhibitor. (E) HMGB1 mRNA expression in 34 RB tissues and 11 normal retina tissues was analyzed by RT-qPCR. *P<0.05 compared with normal retina tissues. (F) HMGB1 mRNA expression in the high miR-665 expression group was significantly lower than that in the low miR-665 expression group. *P<0.05 compared with low miR-665 expression group. (F) Correlation analysis of the expression levels of miR-665 and HMGB1 in RB tissues was performed using Spearman’s correlation analysis. R2=0.3561 P=0.0002.