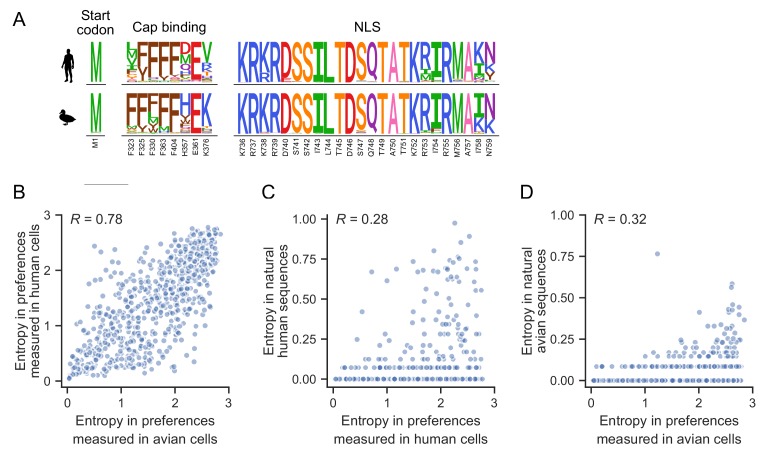
Figure 2. Functional constraints on PB2.
(A) The amino acid preferences measured in human and avian cells for key regions of PB2: the start codon, sites involved in cap-binding, and sites comprising the nuclear localization sequence (NLS). The height of each letter is proportional to the preference for that amino acid at that site. Known critical amino acids are generally strongly preferred in both cell types. (B) Correlation of the site entropy of the amino-acid preferences measured in each cell type. (C) Sites of high variability (as measured by entropy) in natural human influenza sequences occur at sites of high entropy as experimentally measured in human cells. (D) Sites with high variability in natural avian influenza sequences occur at sites of high entropy as experimentally measured in duck cells. Figure 2—figure supplement 1 shows the complete map of amino acid preferences as measured in human and avian cells. Preferences (as well as mutation effect and differential selection for all mutations as calculated for Figure 3) are in Figure 2—source data 1.