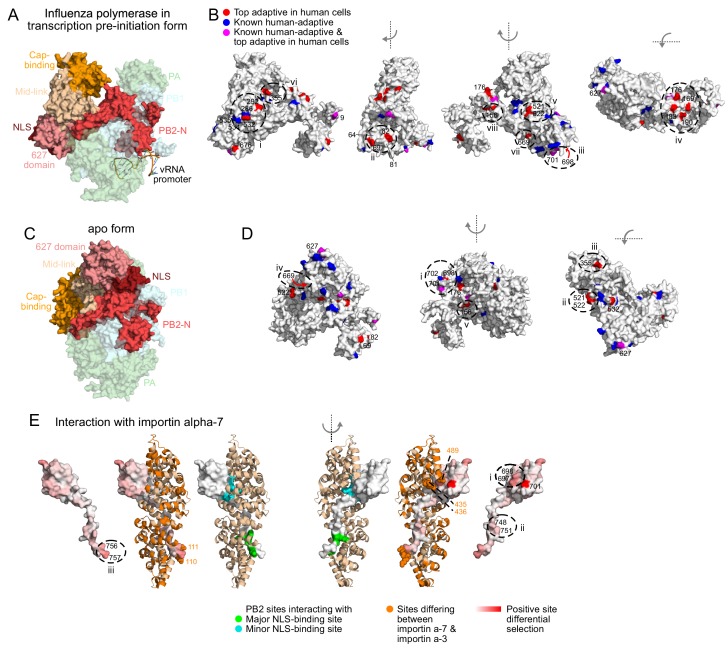
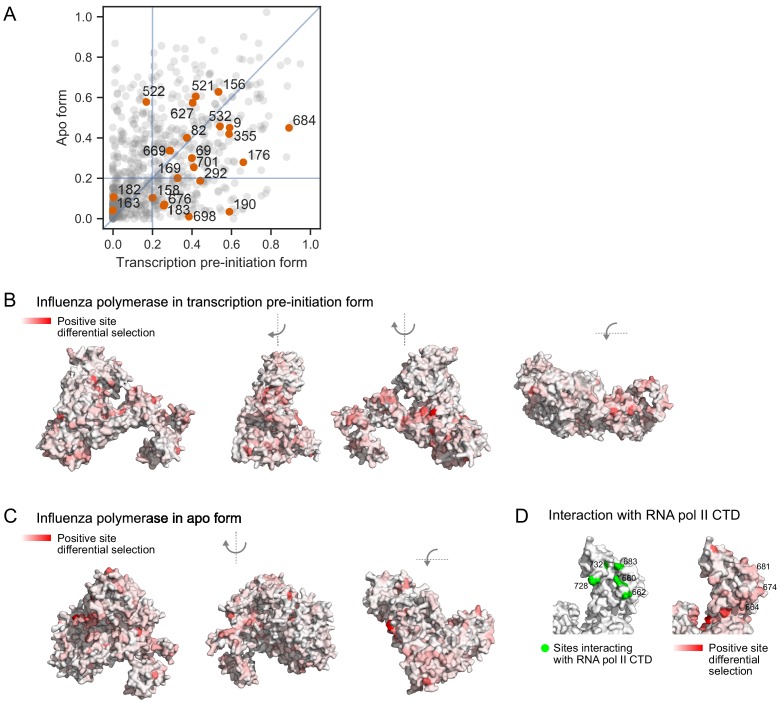
Figure 5. Locations of top human-adaptive mutations on the structure of the influenza polymerase.
Overall structure of the influenza polymerase complex comprising PB2, PB1 and PA in (A, B) the transcription pre-initiation form (PDB: 4WSB) and (C, D) the apo form (PDB: 5D98). PB2 domains defined as in Pflug et al. (2017). (B, D) Sites of top human-adaptive mutations identified by deep mutational scanning are shown in red on the PB2 subunit of the structure. Sites of previously experimentally verified human-adaptive mutations are in blue (25 sites as listed in Figure 3—source data 1). Sites identified by deep mutational scanning and which were also previously known are in purple. A subset of sites are labeled and/or circled for referencing in the main text, to indicate surfaces that might mediate host-interactions. Similar results are obtained if we instead analyze the structures in terms of a continuous variable representing the extent of human-specific adaptation at each site (Figure 5—figure supplement 1B, C). (E) Structure of PB2 C-terminal fragment co-crystalized with importin-α7 (PDB: 4UAD). Sites on PB2 interacting with major and minor NLS binding surfaces of importin-α7 are in green and cyan respectively. Importin-α7 is depicted in ribbon form in tan. We used the deep mutational scanning to define a continuous variable indicating the extent of host-specific adaptation at each site of PB2. Specifically, for each site, we computed the positive site differential selection by summing all positive mutation differential selection values at that site (i.e., the total height of the letter stack in the positive direction in logoplots such as in Figure 3D). We mapped this differential selection onto the PB2 C-terminal fragment in red; PB2 sites with high differential selection are numbered. Regions of importin-α7 that differ from importin-α3 are colored in orange, those near PB2 sites with high differential selection are shown as spheres. For all structures, the avian influenza (S009) PB2 amino acid sequence was mapped onto the PB2 chain by one-2-one threading using Phyre2 (Kelley et al., 2015) (Confidence in models for 4WSB, 5D98, and 4UAD are 100%, 100%, and 99% respectively). Sites are numbered according to the S009 PB2 sequence. Figure 5—figure supplement 1 shows relative solvent accessibility of human-adaptive mutations, as well as positive site differential selection mapped onto structures of influenza polymerase.