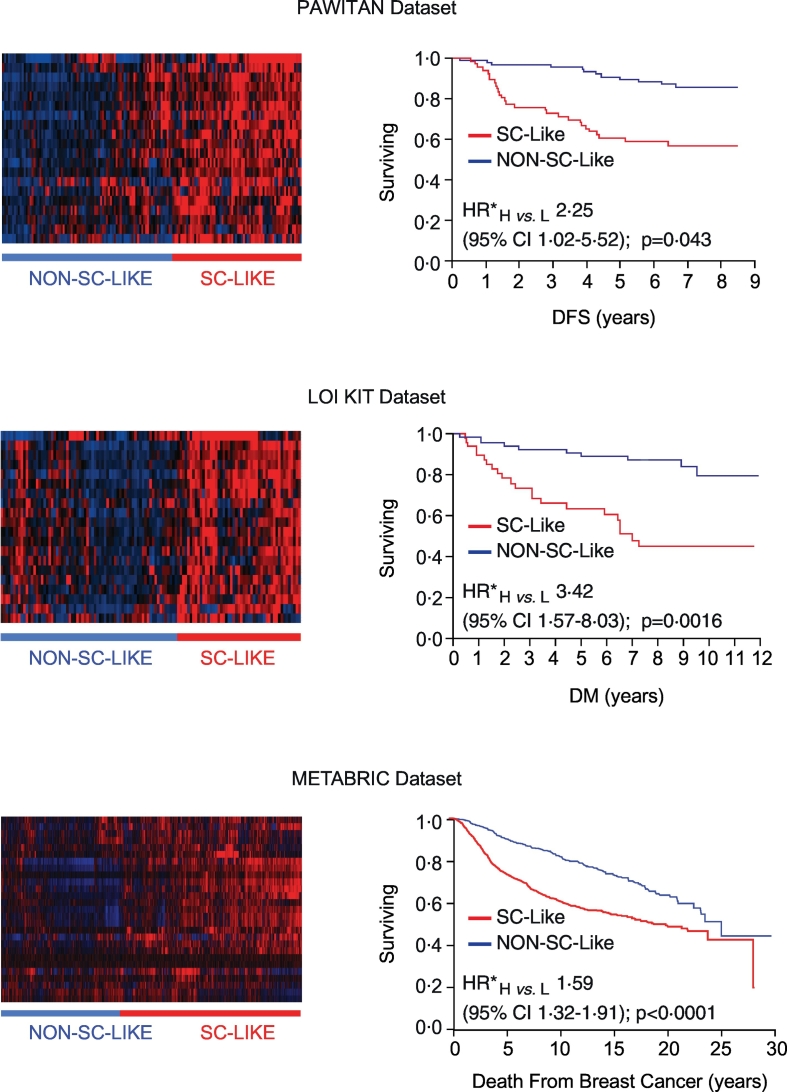
Fig. 2.
Analytical validation of the 20-gene SC signature in independent BC datasets.
Left panels, Identification of “SC-like” and “Non-SC-like” subgroups of BCs by unsupervised hierarchical clustering analysis of the expression of the 20-gene SC signature in the BC datasets published by Pawitan et al. [10] (endpoint: disease-free survival, DFS), Loi et al. [11] (endpoint: time to distant metastasis, DM) and in the METABRIC dataset [12] (endpoint: death from breast cancer). Right panels, Prognostic significance of the “SC-like” (H, for High similarity to SCs) vs. “Non-SC-like” (L, for Low similarity to SCs) classification determined by Kaplan-Meier analysis in the same BC datasets using the endpoints specified above. Multivariable Cox proportional hazards model (see Supplementary Table S2 for details on the multivariable analyses in the different datasets). HR*, multivariable hazard ratio; CI, confidence interval; p, p-value.