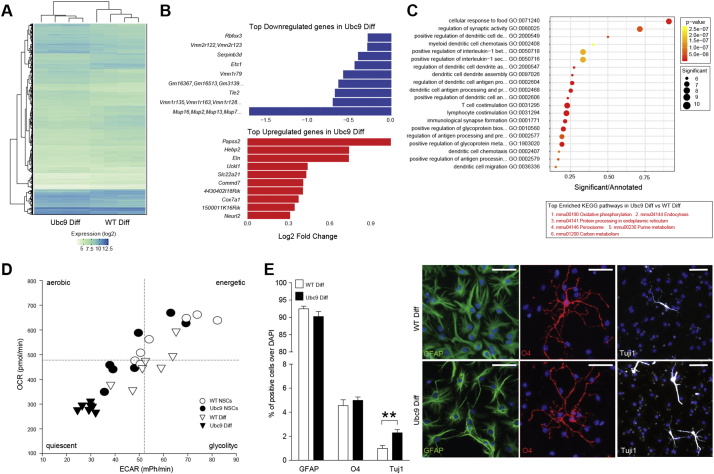
Fig. 2.
Ubc9 NSCs show a quiescent metabolic profile upon differentiation and are predisposed to neuronal differentiation in vitro. (A) Heatmap of genes differentially expressed in Ubc9 Diff vs WT Diff cells. N = 4 per group, adjusted p-value<0.05. See also Data S2. (B) Bar chart of the fold change (log2) in Ubc9 Diff vs WT Diff cells of the 10 most upregulated and downregulated genes. All genes have an adjusted p-value<0.05. See also Data S2. (C) Plot of the GO enrichment results for genes differentially expressed in Ubc9 Diff vs WT Diff cells. The x-axis shows the enrichment ratio, the color indicates the enrichment p-value (Kolmogorov–Smirnov statistic), and the size of each point indicates the number of significant genes in the corresponding GO category. The table at the bottom shows the 6 KEGG pathways showing the strongest enrichment (GAGE analysis p-value<0.05 for all pathways, see methods) in the same comparison. See also Data S2. (D) OCR vs ECAR graph of undifferentiated and differentiated WT and Ubc9 NSCs. Data are expressed as mean pmoles/min (OCR) and mean milli-pH units (mpH)/min (ECAR) ± SEM. N = 7 per group from 2 independent experiments.
(E) Quantification and representative immunofluorescence-staining of WT Diff and Ubc9 Diff NSCs. Data are means ± SEM. N ≥ 6 per group from 2 independent experiments, **p-value<0.01 (2-tailed Student's t-test). Nuclei are stained with DAPI, scale bars: 50 μm.