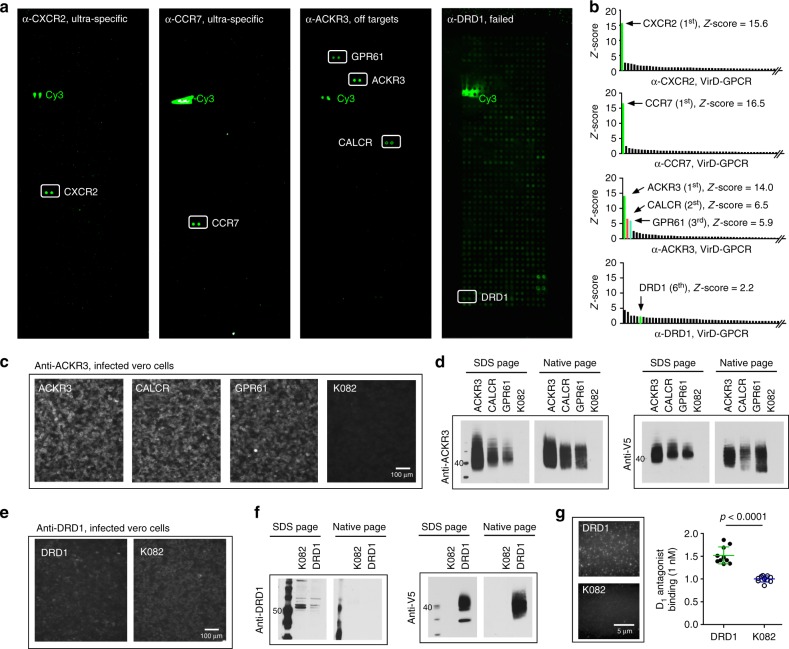
Fig. 2.
Specificity tests of commercial mAbs on VirD-GPCR arrays. a Examples of binding signals obtained with commercial mAbs. Anti-CXCR2 and -CCR7 are shown as ultra-specific; anti-ACKR3 can cross-react with CALCR and GPR61; anti-DRD1 completely failed to recognize its target while showing nonspecific binding activities to other GPCRs. b Histograms of Z-scores obtained with three mAbs. Z-scores of the two off- targets identified by anti-ACKR3 are also shown. c Immunofluorescence analysis (IFA) validation of anti-ACKR3 to its off-targets in infected Vero cells. K082-infected cells are shown as a negative control. d Immunoblot analysis also confirmed that anti-ACKR3 can recognize its two off-targets in the cell lysates of infected Vero cells under both denatures and native conditions. e, f Anti-DRD1 failed to recognize DRD1 in VirD-DRD1-infected cells using IFA (e) or immunoblot (IB) analyses (f). g Single-molecule imaging using TIRF microscopy to determine interactions between VirD-DRD1 and its canonical ligand D1 antagonist. K082 virions were used as a negative control. Quantitative analysis of TIRF imaging demonstrated that VirD-DRD1 showed significantly higher binding signals to D1 antagonist than K082. Data were analyzed with two-tailed Student’s t-test, n = 10