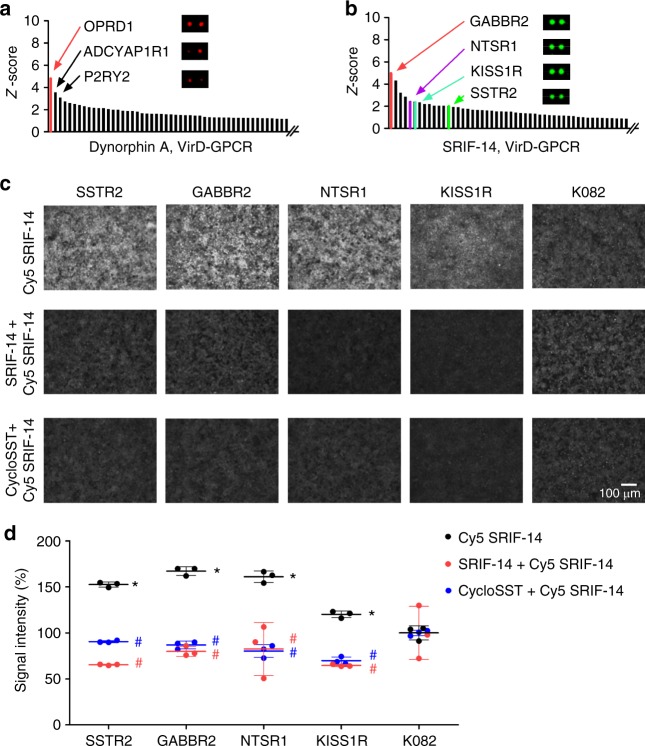
Fig. 3.
Identification and cell-based validation of peptide ligand–GPCR interactions. a Commercially available Dynorphin A was Cy5-labeled and probed to a VirD-GPCR array. Quantitative analysis showed that it bound to VirD-OPRD1 with the highest Z-score followed by ADCYAP1R1 and P2RY2. b A commercially available peptide SRIF-14 was Cy3-labeled and probed to a VirD-GPCR array. Quantitative analysis revealed that it bound to several unexpected off-targets in addition to its canonical receptor, SSTR2. c Vero cells were separately infected with SSTR2, GABBR2, NTSR1, KISS1R, or K082 virus. Infected cells were then incubated with Cy5-labeled SRIF-14 at 8 µM in the absence (upper panel) or presence of cold SRIF-14 (middle panel) or cyclosomatostatin (cycloSST, lower panel). d Quantitative analysis of binding signals. Each binding assay was performed in triplicate and the obtained binding signals were normalized to those of the K082 controls. Data were analyzed by two-way ANOVA with repeated measures followed by Bonferroni post-test. *P < 0.05, comparison between VirD-GPCRs and K082 in the absence of competitor ligands; #P < 0.001, comparison between binding signals obtained in the absence and presence of the competitor ligands. n = 3, biologically independent samples