Fig. 2.
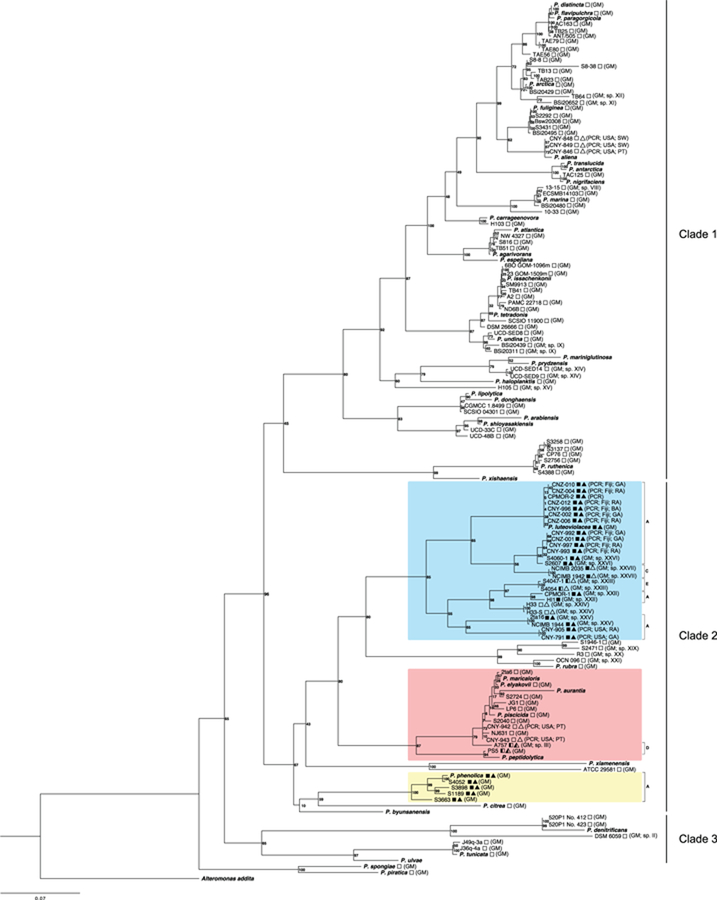
Pseudoalteromonas species phylogeny and bmp gene cluster distribution. A maximum likelihood phylogeny generated from concatenated 16S, gyrB, pyrH, recA and rpoD nucleotide sequences. The three least inclusive clades containing the bmp cluster are highlighted in color. Lettered brackets (A, C, D and E) indicate the version of the bmp cluster. The 42 type strains are indicated in bold italics and the 17 strains isolated as part of this study by CN prefixes followed by country of origin (USA or Fiji) and source (SW = seawater, PT = plankton tow, GA = green alga, RA = red alga, BA = brown alga). Squares indicate bmp distribution (filled = present, empty = absent) and bmp version (fully filled = version A, half-filled = partial BCG). Method of detection (GM = genome sequence or PCR) indicated. Filled triangles indicate the detection of pentabromopseudilin, half-filled triangles indicate the detection of tetrabromopyrrole but not pentabromopseudolin and empty triangles indicate that no compounds associated with the bmp gene cluster were detected. Roman numerals (as designated in Fig. 3) indicate 17 candidate new species identified based on coherence between 95% ANI groupings and the species phylogeny.
