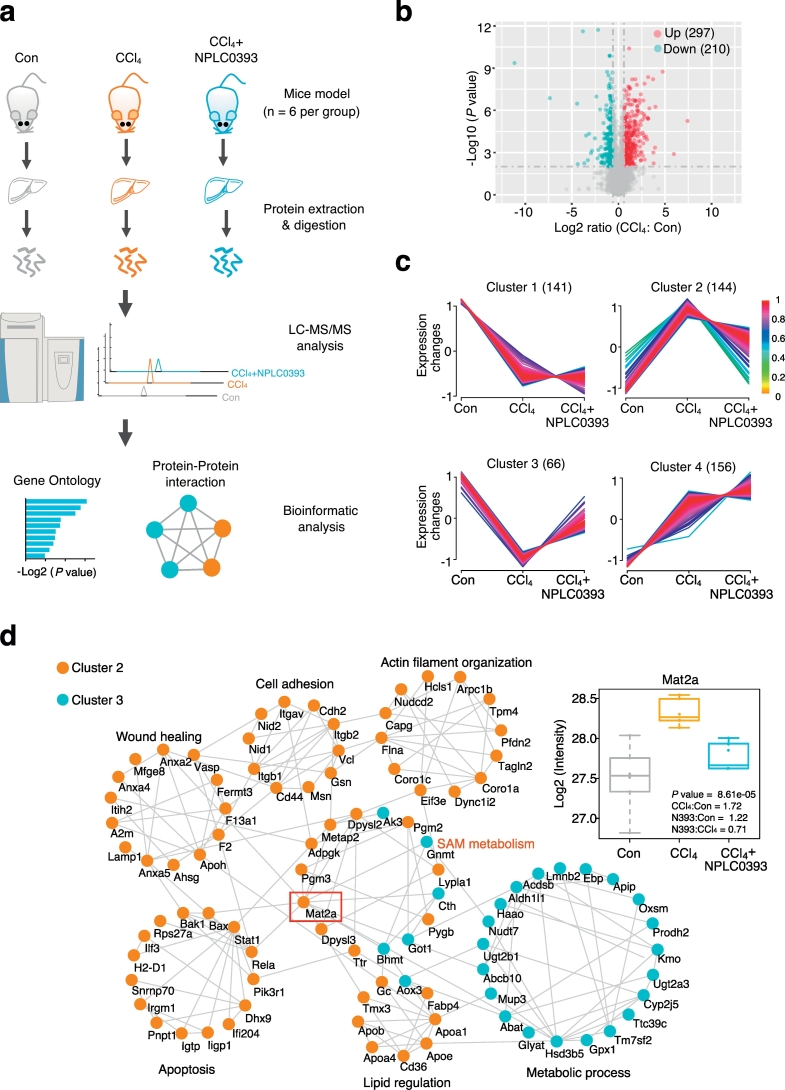
Fig. 1.
pharmacoproteomic analysis of the mice livers from control, fibrosis model and NPLC0393 treatment groups. (a) Workflow chart of label-free quantitative proteomic experiment. Total protein was extracted from different mice liver tissues (n = 6 for Control, CCl4 and NPLC0393 groups respectively) and digested with trypsin. Equal amount of resolved peptides were analyzed by LC-MS/MS. Bioinformatic analysis of differentially expressed proteins was carried out using STRING database. (b) Volcano plot illustrating proteins with different abundance in CCl4 and Control groups. The Log2 ratios of protein intensities of CCl4 to Control group were plotted against the negative Log10 P values. Red points represent upregulated proteins in CCl4 group, blue points represent downregulated proteins in CCl4 group (fold change >1.5, P value <.01) and gray points represent unchanged proteins. (c) Clustering of the differentially expressed proteins. The changed proteins were assigned to four clusters by fuzzy c-means (FCM) clustering algorithm. The y axis is log10 transformed and normalized, and the number of proteins in each cluster is indicated in parentheses. Each trace was colored according to its membership value in the corresponding cluster (referred to membership colour bar). (d) Protein-protein interaction analysis of proteins in Cluster 2 and Cluster 3 using STRING database. Interactions between two proteins were indicated with gray edges. Colour of the node indicates clusters (Orange: Cluster 2; Blue: Cluster 3; unconnected proteins were not shown). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)