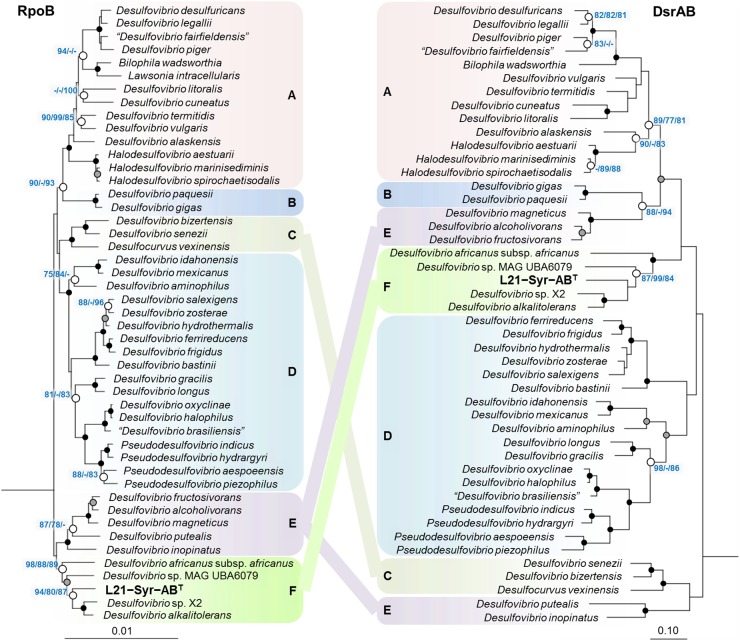
FIGURE 2.
Placement of strain L21-Syr-ABT within the family Desulfovibrionaceae based on phylogenies of complete RpoB and concatenated DsrAB protein sequences. Tree topologies were reconstructed under the maximum-likelihood criterion and rooted using the respective sequences of Desulfonatronum lacustre, Desulfonatronospira thiodismutans, Desulfonatronovibrio hydrogenovorans, Desulfovermiculus halophilus, Desulfohalobium retbaense, Desulfothermus naphthae, Desulfonauticus submarinus, Desulfoplanes formicivorans, Desulfomicrobium baculatum, and Desulfobacter postgatei as outgroup (not shown). Strains and accession numbers of genomes used for the extraction of protein sequences are provided in Supplementary Table S1. Note, that in Bilophila wadsworthia the dsrB and dsrD genes are combined, but for tree reconstruction the sequence of the DsrD protein was removed. Support of a distinct branching by bootstrap analyses is indicated by symbols (see caption of Figure 1). Boxes shaded in different colors indicate distinct clades supported by significant bootstrap values in the RpoB tree. Scale bars indicate changes per amino acid position.