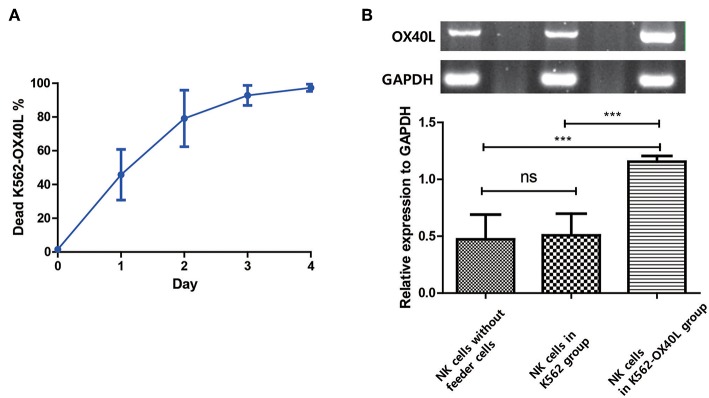
Figure 7.
OX40L on NK cells is generated after exposure to K562-OX40L and maintained during NK cell expansion (A) Survival of K562-OX40L feeder cells during NK cell expansion. γ-irradiated K562-OX40L were stained with 5μM CFSE prior to co-culturing with PBMCs. Then, CFSE-positive K562 and PI-stained dead cells were identified every day by flow cytometry. n = 3 error bars, mean ± SD (B) OX40L mRNA expression in NK cells. NK cells were isolated by using NK cell enrichment kit at day 3 from PBMCs without feeder cells, PBMCs with either K562 or K562-OX40L culture conditions. Then, RT-PCR with GAPDH serving as control was performed. Band intensity from RT-PCR results were quantified using Image J after color inversion of the image. OX40L gene expression relative to GAPDH expression was analyzed from triplicate samples of each group. (n = 3 error bars, mean ± SD; ***p < 0.001).