Figure 7.
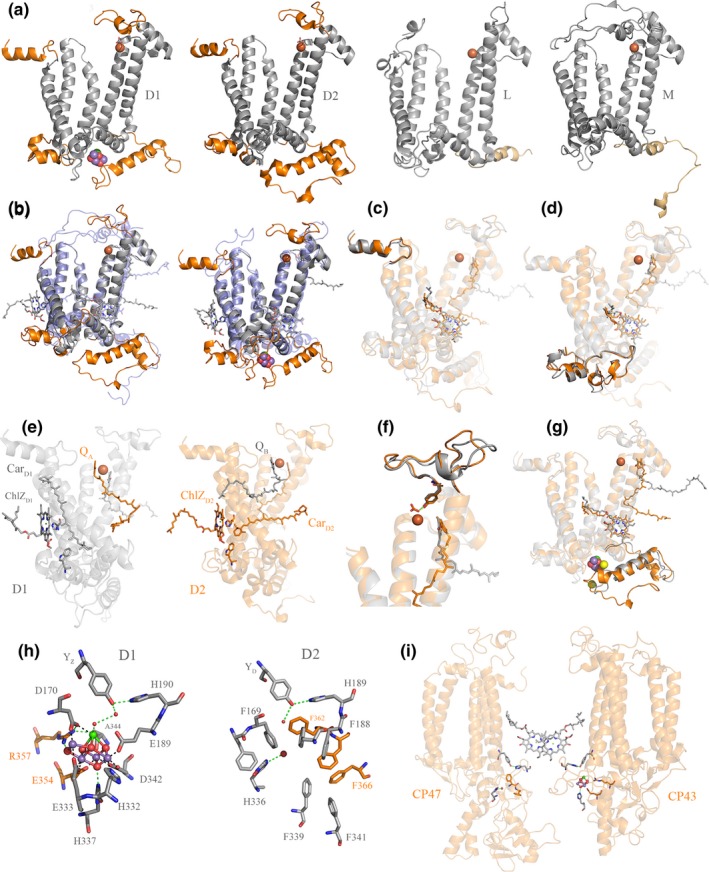
Structural comparisons of Type II reaction center proteins. (a) Several structural domains are conserved in D1 and D2 but are absent in L and M: These are highlighted in orange. D1 and D2 are plotted from the crystal structure of PSII from Thermosynechococcus vulcanus, PDB: 3WU2 (Umena et al., 2011) and L and M from Thermochromatium tepidum, PDB: 3WMM (Niwa et al., 2014). (b) Overlap of D2 (gray) with M (transparent blue) and D1 (gray) with L (transparent blue). (c) Overlap of D1 (gray) and D2 (orange) highlighting the conserved N‐terminus. (d) Overlap of D1 and D2 highlighting the conserved protein fold between the 1st and 2nd transmembrane helices. (e) D1 is shown in gray and D2 in orange. ChlZD 1, CarD1, W105, and QB are shown in gray sticks. ChlZD 2, CarD2, W104, and QA are shown in orange sticks. (f) Overlap of D1 and D2 highlighting the conserved protein fold where the bicarbonate binding site is placed. (g) Overlap of D1 and D2 highlighting the conserved protein fold at the C‐terminus. (h) The Mn4CaO5 cluster coordination sphere and the equivalent location in D2. (i) Perspective view showing the interaction of CP47 and CP43 with the electron donor side of D2 and D1, respectively. The structural homology of CP43 and CP47 indicates that these originated from a gene duplication event making the homodimeric antenna to a homodimeric core (Cardona, 2016). The reason why CP47, and in particular CP43, interact with the donor side of PSII is an unsolved mystery given the fact that their main role is that of light harvesting. It can be rationalized however if water oxidation started in a homodimeric reaction center early during the evolution of photosynthesis (Cardona, 2017) [Colour figure can be viewed at wileyonlinelibrary.com]
