Figure 2: Perils of cancer model evolution.
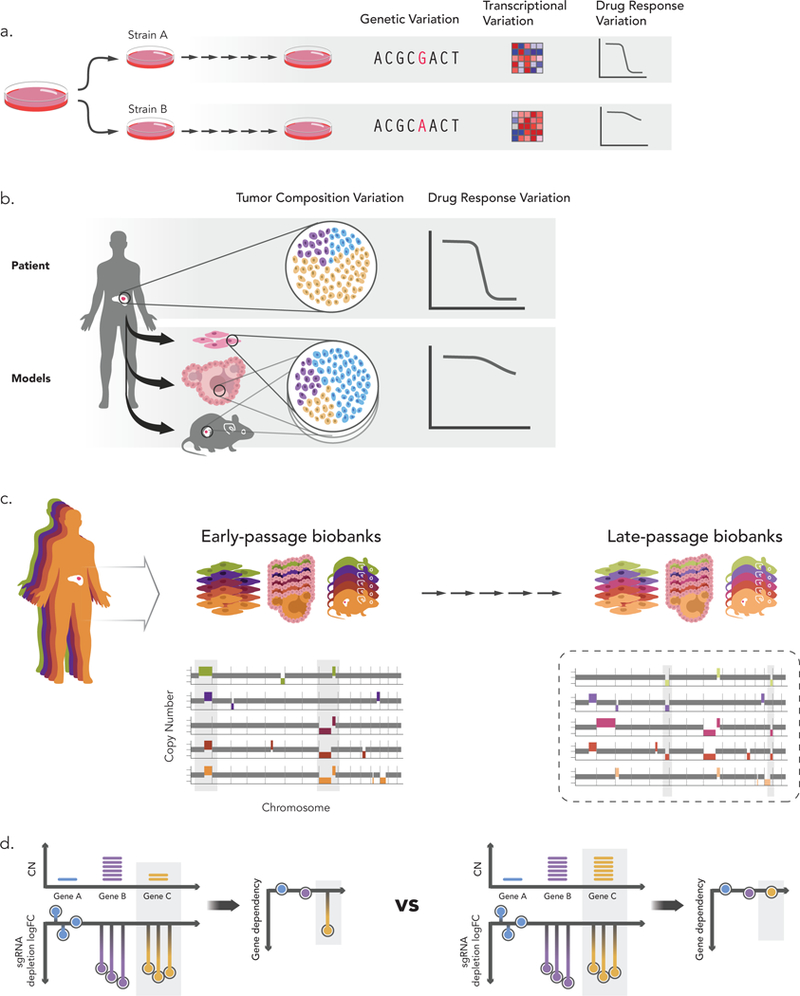
(a) Genomic evolution leads to variability across strains of the same model. Genetic alterations acquired during model propagation can translate into differential gene expression patterns and result in disparate drug response68. (b) Cancer models are used as tumor “avatars” to predict drug response in the patient from which the model was derived. However, if a rare subclone expands in the model and becomes dominant, its drug response may not reflect that of the primary tumor81. (c) Cancer models are also used as cohorts, also known as biobanks, to characterize genotype-phenotype relationships (e.g., cancer dependencies associated with specific mutations). Models are usually characterized upon their derivation, but functional experiments keep being conducted throughout model life. Continued passaging of the models may alter their genomic profiles, thus confounding analyses that make use of early-passage genomic profiles and late-passage functional experiments81. (d) CRISPR/Cas9 screen results need to be corrected for copy-number to account for the number of cuts (a phenomenon known as the CRISPR copy-number effect). Changes in aneuploidy (and other copy-number alterations) as a result of genomic evolution can jeopardize the accuracy of this computational correction. For example, if an amplification of gene C occurred between genomic characterization and CRISPR screening, and the original genomic characterization is used for correcting the copy-number effect, this gene could be mis-identified as a genetic dependency.
