Figure 3: New research opportunities presented by cancer model evolution.
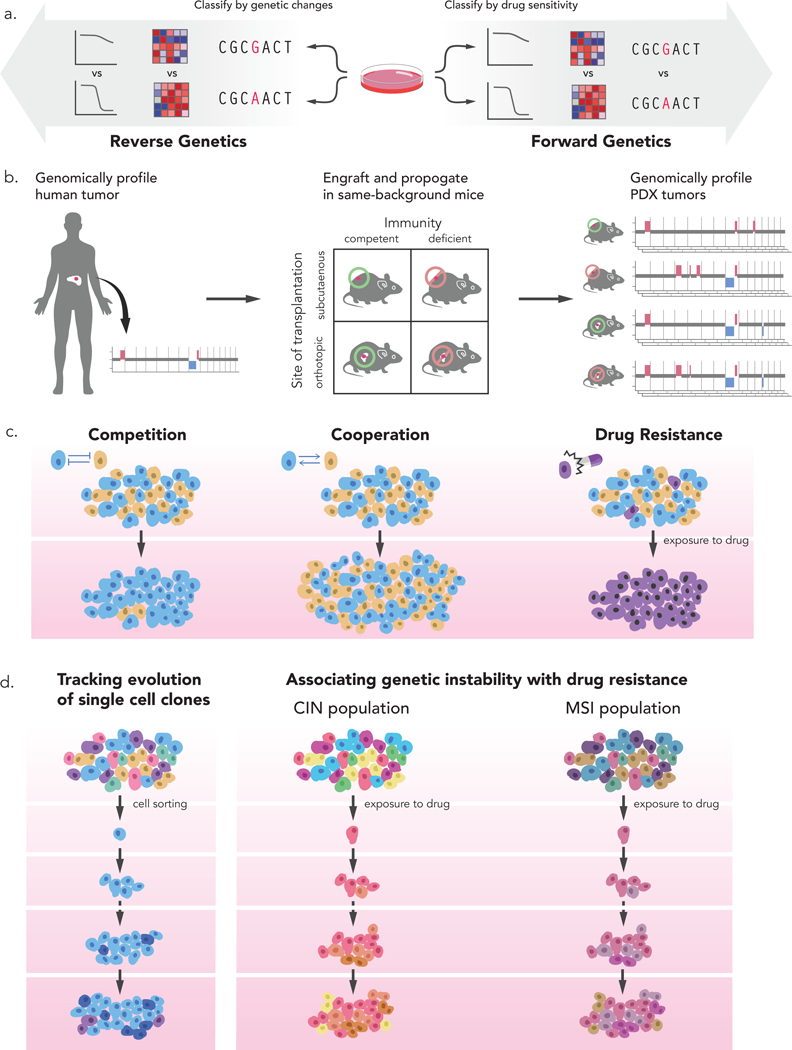
(a) The natural genomic evolution of cancer models generates semi-isogenic model strains that can be used both for reverse genetics and for forward genetics. In reverse genetics experiments, the gene expression profiles and drug response patterns of model strains with a genetic alteration of interest can be compared to those of strains without that alteration. In forward genetics experiments, the genetic landscapes and gene expression profiles of model strains that are sensitive to a drug of interest can be compared to those of strains that are resistant to that drug. (b) Genomic evolution can be used to study the selection pressures that shape the genetic landscapes of tumors. For example, PDXs can be generated in mice hosts that share their genetic background, but differ in their immune status and transplantation sites. The genomic profiles of these PDX models following propagation can be characterized and compared to that of the primary human tumor. The rate, extent and identity of genomic alterations in the various models can help identify the components that determine the evolutionary trajectory of tumorigenesis. (c) Cancer model evolution can be used to study cancer heterogeneity and cellular interactions. Competitive and cooperative interactions between tumor subclones can be dissected using single-cell genomics, genome editing and cell barcoding technologies. Drug sensitivity and resistance can also be studied by following tumor evolution and clonal dynamics following drug exposure. (d) Mechanisms of genomic instability itself can be studied by following how single-cell clones become heterogeneous (left), or by following how drug resistance mechanisms differ between models that harbor distinct deficiencies in genome maintenance pathways (right). CIN, chromosomal instability; MSI, Microsatellite instability.
