Figure 5.
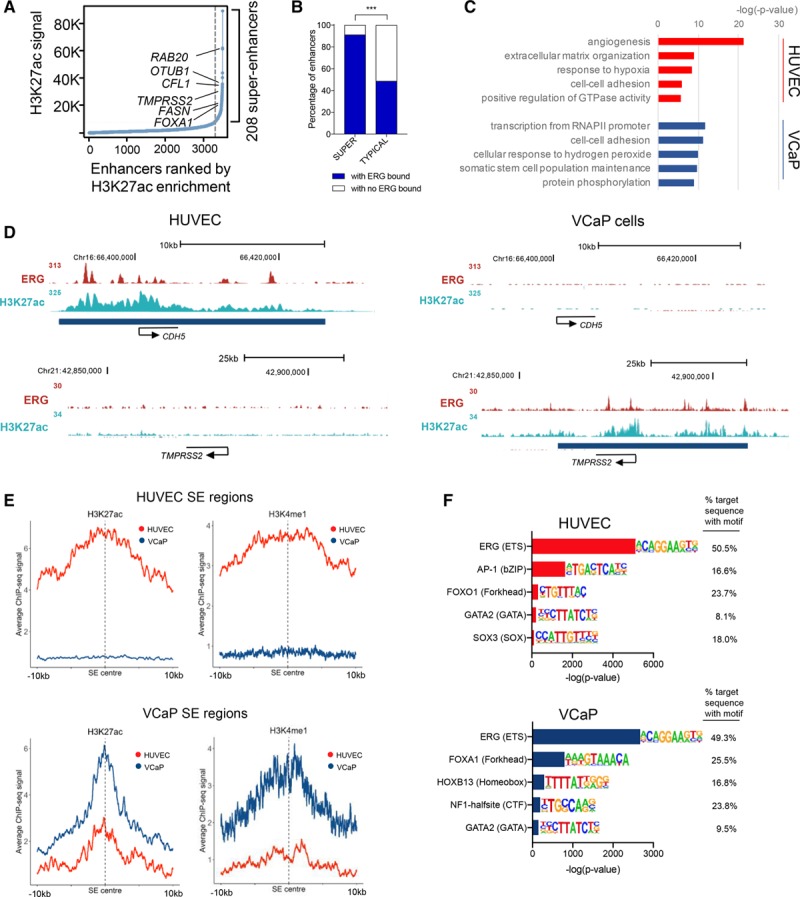
ERG (ETS-related gene) associates with lineage-specific super-enhancers (SEs). A, Identification of human prostate epithelial cancer cell line (VCaP) SE by H3K27ac chromatin immunoprecipitation with deep sequencing (ChIP-seq) signal in rpm/bp (see also Online Table VII). B, ERG binding at SE and typical enhancers in VCaP cells. SE have significantly more ERG binding than typical enhancers; ***P<0.0001, Fisher exact test. C, Gene ontology analysis on the SE-associated genes in human umbilical vein endothelial cell (HUVEC; red) and VCaP cells (blue). The top 5 biological processes in both cells are ranked by significance (−log (P value)). D, ChIP-seq binding profiles for ERG and H3K27ac occupancy in HUVEC (left) and VCaP cells (right). H3K27ac-defined SE regions are depicted as blue bars below tracks. E, Aggregate plots of active histone modifications H3K27ac and H3K4me1 histone modifications from HUVEC and VCaP cells in HUVEC SE (top) and VCaP SE (bottom). Plots centered on the SE center. The average size of VCaP SE is smaller than those identified in HUVEC. F, Motif analysis at ERG-binding sites (±200 bp) in HUVEC and VCaP. Top 5 most enriched transcription factor families are shown with the most highly occurring member of each family represented. Motifs for the binding of critical lineage transcription factors coincide with the ERG motif in both HUVEC and VCaP cells.
