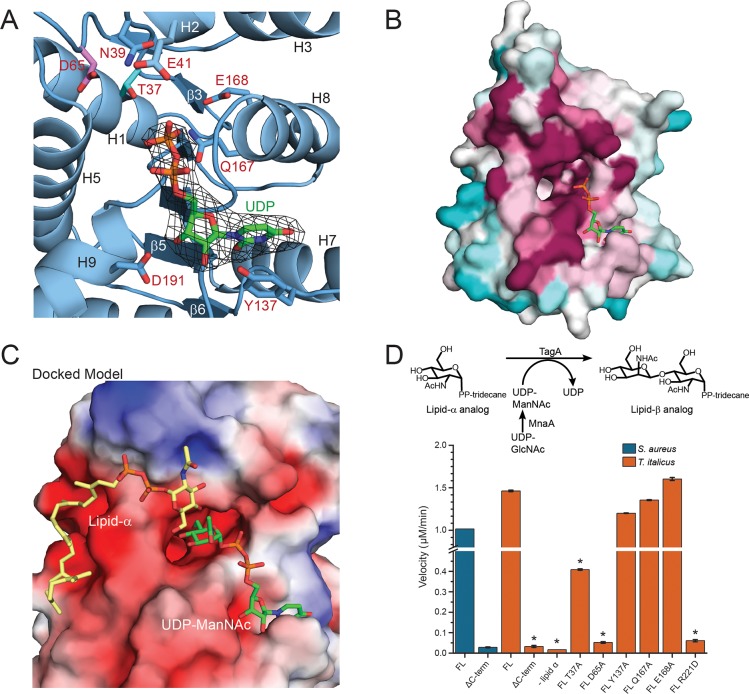
Fig 2. In silico substrate binding and mechanistic studies of TagA.
(A) Structure of the UDP:TagAΔC complex. A simulated annealing omit map contoured at 3σ reveals that the β-phosphate of UDP (orange) in the UDP:TagAΔC structure is adjacent to the proposed catalytic base, Asp65 (pink), and putative ManNAc stabilizing residue, Thr37 (cyan). The uracil nucleoside is pi-stacked over Tyr137. (B) Consurf analysis reveals that UDP projects its phosphates (orange) into the pocket and is coordinated by a pocket of highly conserved residues. Highly conserved (magenta), moderately conserved (white) and weakly conserved (teal) residues are indicated. (C) In silico generated model of TagAΔC bound to its substrates. The model contains a lipid-α analog (yellow) and UDP-ManNAc (green). The electrostatic surface of the protein is shown. Negatively charged residues (red), neutral residues (white), positively charged residues (blue) are shown. The coordinates of the model were generated using a two-step procedure. First, the coordinates of the ligand-bound UDP:TagAΔC crystal structure were used to restrain the positioning of UDP-ManNAc. Autodock vina was then used to dock lipid-α. The docking results positioned the non-reducing end of GlcNAc toward the C4 in ManNAc when bound to the TagAΔC dimer. (D) The upper panel indicates the reaction scheme used for the in vitro TagA activity assay. 200 nM TagA enzyme was incubated at 30°C with 100 μM lipid-α substrate analog and UDP-ManNAc produced in situ from UDP-GlcNAc by the epimerase MnaA, followed by quenching with 4M urea. Conversion of UDP-ManNAc to UDP is monitored at 271 nm using a DNAPak PA200 anion exchange column. The lower panel indicates activity measurements of T. italicus and S. aureus TagA enzymes from the in vitro TagA activity assay, as described above. Reactions with error bars were performed in triplicate, and asterisks indicate p<0.005 by Student’s T-test.