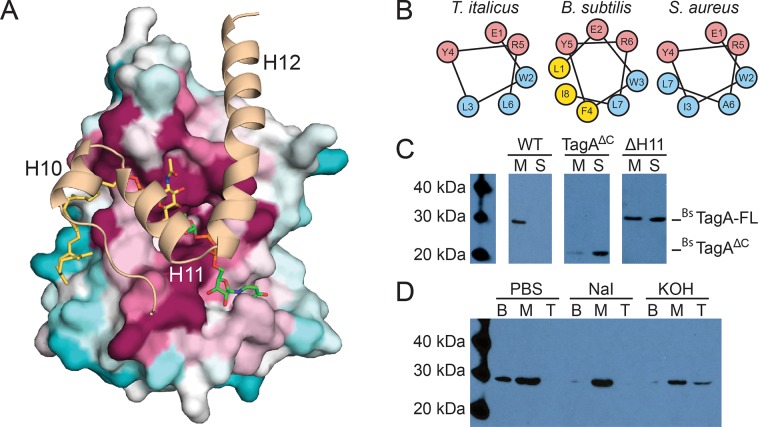
Fig 3. Computational and biochemical studies of the TagA enzyme inform cellular localization.
(A) A model of full-length TagA was constructed with experimentally determined TagAΔC (surface representation) and the C-terminal domain, which was modeled by GREMLIN structural prediction (cartoon representation). Three C-terminal helices appear to complete the active site and obstruct the dimeric interface of TagAΔC. Highly conserved (magenta), moderately conserved (white) and weakly conserved (teal) residues are indicated for the TagAΔC crystal structure. (B) Helical wheel projections of helix H11 in TagA homologs predict a putative amphipathic helix. (C) TagA associates with the bacterial cell membrane. Immunoblots of cellular fractionation indicate that B. subtilis TagA is exclusively localized to the membrane (M), while TagAΔC is primarily localized in the supernatant (S). Samples were fractionated by ultracentrifugation identically and the BsTagA-FL blot was exposed for 10 minutes, the BsTagA-V196 blot was exposed for 1 minute, and the BsTagA-ΔH11 blot was exposed for 30 seconds. (D) TagA is a peripheral membrane protein. Chaotropic and alkaline treatments of B. subtilis TagA reveal that the enzyme is peripherally associated with the membrane and is more effectively displaced by alkaline treatment. Treated membrane fractions were loaded onto a sucrose cushion, centrifuged, and carefully separated into bottom (B; pellet), middle (M; sucrose cushion volume), and top (T; sample volume).