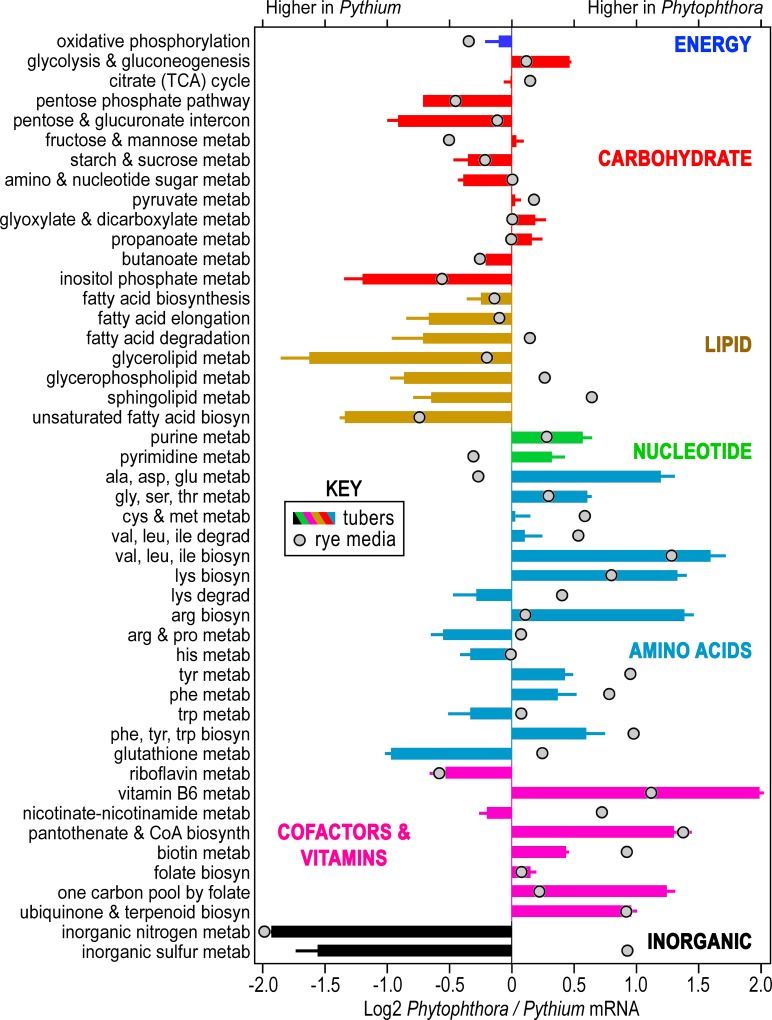
Fig 2. Comparison of expression of metabolic genes during tuber infection and growth on artificial media by Ph. infestans and Py. ultimum.
Graphed is the log2 ratio of the summed FKPM of genes for each species. Genes were grouped based on pathway maps in the KEGG database, except for genes involved in the assimilation of inorganic nitrogen and sulfur which are listed separately. Bars represent the ratio of expression in the two species at the early infected tuber timepoint, with bars pointing to the right representing higher expression in Ph. infestans. Each bar is color-coded by category (e.g. carbohydrate, lipid, nucleotide, amino acid cofactor/vitamin, and inorganic sulfur or nitrogen metabolism). Error bars are based on three biological replicates. Circles represent the expression ratios in the early rye media timepoint.