Mononuclear phagocytes comprise monocytes, macrophages, and dendritic cells (DCs) with several tissue-resident subsets described according to surface markers, function, and ontogeny.1 Macrophages alone are entirely tissue-resident, do not circulate, and form a network of immune sentinels in all organs, poised to detect a range of perturbations, from infection to tissue damage. Although initially thought to be derived from hematopoietic stem cells via monocyte differentiation, recent fate-mapping studies in mice have established that some tissue macrophages are seeded prenatally, and may originate from yolk-sac or fetal liver progenitors, with the proportion of monocyte-derived or prenatally seeded macrophages varying in different organs.1 Regardless of ontogeny, murine studies suggest that macrophages are profoundly influenced by their tissue environment and take on organ-specific phenotypes and transcriptional signatures influenced by the local cell composition and by sampling the milieu.2
Why should nephrologists care about macrophages? Several studies show that, at least in mice, tissue macrophages can play an important role in organ homeostasis; for example, in the heart, macrophages are intimately associated with the conducting system and are required for normal electrical conduction.3 In the gut, macrophages interact with enteric neurons to promote peristalsis.4 Murine data also suggest that kidney macrophages are likely to modulate susceptibility to, and the resolution of, kidney infection, ischemia-reperfusion injury, GN, diabetic nephropathy, and progression to fibrosis.5 Notably, the methods used in these studies largely do not distinguish between monocyte-derived or embryonically seeded macrophages, often affect macrophages and DCs, and utilize global rather than kidney-specific macrophage depletion. However, even with these caveats, the overall body of evidence suggests that resident kidney macrophages are important players in injury responses, with both positive and negative effects.6
One of the problems of translating mouse studies of kidney macrophages to humans is that some of the markers used to delineate mouse macrophages are not expressed by their human counterparts, making it difficult to identify analogous populations across specifies. For example, the F4/80 antibody (binding to Adgre1) is mouse macrophage–specific. Further, it has emerged that markers that were historically considered to distinguish DCs from macrophages such as CD11c and MHCII, are frequently expressed by tissue macrophages. Indeed, monocyte-derived macrophages in human skin and kidney are CD11c+/MHCII+/CD14+.7,8 In clinical histopathology, CD68 is often used as a macrophage marker in human kidney, and CD206 expression has also been described on human kidney macrophages, but the specificity of these markers is unclear. This highlights the problem that Zimmerman et al.9 and colleagues attempt to address: is it possible to identify a cross-species kidney macrophage–specific “signature” or marker set that will facilitate the translation of murine models to human biology and disease?
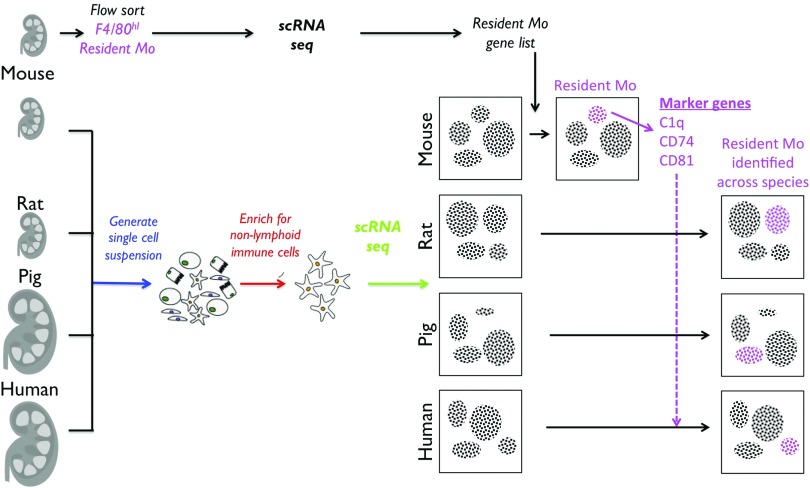
To tackle this problem, they turned to single-cell RNA sequencing (scRNAseq). To date, transcriptomic studies of immune cells have largely relied on the measurement of mRNA in subsets isolated by flow sorting or magnetic bead selection, requiring prior knowledge of subset-specific markers. However, advances in single-cell technologies10 now enable immune cells to be profiled in an unbiased manner, such that those with a similar transcriptome can be clustered without any preconceived idea about their canonical markers. Zimmerman et al.9 leveraged this strength in their study. They isolated kidney CD45+ (immune) cells from one mouse, rat, pig, and human, and performed scRNAseq on a mononuclear phagocyte–enriched cell suspension. Using the data generated, they clustered cells according to transcriptional similarity, allowing them to identify putative “monocyte-derived” and “resident” kidney macrophage cell clusters, and confirmed overlap with the transcriptional signatures obtained from flow-sorted F4/80lowCD11bhi and F4/80highCD11blow mouse kidney macrophages, respectively (Figure 1). They used the most differentially expressed gene in the mouse resident macrophage cluster, C1qc, to identify analogous clusters in the human, pig, and rat kidney scRNAseq datasets, and confirmed that the C1q-expressing clusters across species expressed other genes that were highly expressed in mouse macrophages: Cd81, Cd74, and Apoe. To validate these markers, they identified commercially available antibodies for the protein products of three of these genes: C1q, CD74, and CD81. The antibodies were applied to flow cytometric studies in mouse, rat, and human kidneys, and although C1q was not highly expressed in human kidney macrophages, they were able to identify CD74 and CD81 expression on a population of kidney immune cells across species. Finally, they performed a parabiosis experiment in mouse, which suggested that the CD74/CD81+ cells in the kidney were not exchanged, confirming the utility of these markers to identify a bona fide resident macrophage population.
Figure 1.
Summary of experimental setup used by Zimmerman et al.9 to identify a cross-species renal-resident macrophage signature. The resident macrophage cell cluster in mouse kidney (pink) was identified by comparing the gene expression within each cluster to that of sorted resident macrophages (top panel). Marker genes for the resident macrophage population were identified from this mouse cluster and applied to the data obtained from other species, allowing the resident macrophage population to be identified across species.
Of note, this study included cells from only one kidney from each species in their scRNAseq experiment. Because humans are genetically heterogeneous and have variable environmental exposures, the extent to which the proportions of immune cell identified from this single human kidney, and the magnitude of expression of any given marker gene in each cell population, may not be generally applicable. However, even with this caveat, the study demonstrates the power of scRNAseq to identify a core resident macrophage signature as well as novel cross-species kidney macrophage markers that can be validated in future studies, and will facilitate clinically relevant translational research.
Disclosures
None.
Acknowledgments
Dr. Clatworthy is supported by the National Institutes of Health Research (NIHR) Cambridge Biomedical Research Centre, by a Chan Zuckerberg Initiative Human Cell Atlas Technology Development grant, a Medical Research Council New Investigator research grant (MR/N024907/1), an Arthritis Research UK Cure Challenge research grant, 21777, and an NIHR Research Professorship (RP-2017-08-ST2-002).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related article, “Single-Cell RNA Sequencing Identifies Candidate Renal Resident Macrophage Gene Expression Signatures across Species,” on pages 767–781.
References
- 1.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. : Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nat Rev Immunol 14: 571–578, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A-Gonzalez N, Quintana JA, García-Silva S, Mazariegos M, González de la Aleja A, Nicolás-Ávila JA, et al. : Phagocytosis imprints heterogeneity in tissue-resident macrophages. J Exp Med 214: 1281–1296, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hulsmans M, Clauss S, Xiao L, Aguirre AD, King KR, Hanley A, et al.: Macrophages facilitate electrical conduction in the heart. Cell 169: 510–522.e20, 2017. [DOI] [PMC free article] [PubMed]
- 4. De Schepper S, Verheijden S, Aguilera-Lizarraga J, Viola MF, Boesmans W, Stakenborg N, et al.: Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell 175: 400–415.e13, 2018. [DOI] [PubMed]
- 5.Nelson PJ, Rees AJ, Griffin MD, Hughes J, Kurts C, Duffield J: The renal mononuclear phagocytic system. J Am Soc Nephrol 23: 194–203, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers NM, Ferenbach DA, Isenberg JS, Thomson AW, Hughes J: Dendritic cells and macrophages in the kidney: A spectrum of good and evil. Nat Rev Nephrol 10: 625–643, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGovern N, Schlitzer A, Gunawan M, Jardine L, Shin A, Poyner E, et al. : Human dermal CD14+ cells are a transient population of monocyte-derived macrophages. Immunity 41: 465–477, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berry MR, Mathews RJ, Ferdinand JR, Jing C, Loudon KW, Wlodek E, et al.: Renal sodium gradient orchestrates a dynamic antibacterial defense zone. Cell 170: 860–874.e19, 2017. [DOI] [PubMed]
- 9.Zimmerman KA, Bentley MR, Lever JM, Li Z, Crossman DK, Song CJ, et al. : Single-cell RNA sequencing identifies candidate renal resident macrophage gene expression signatures across species. J Am Soc Nephrol 30: 767–781, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potter SS: Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol 14: 479–492, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]