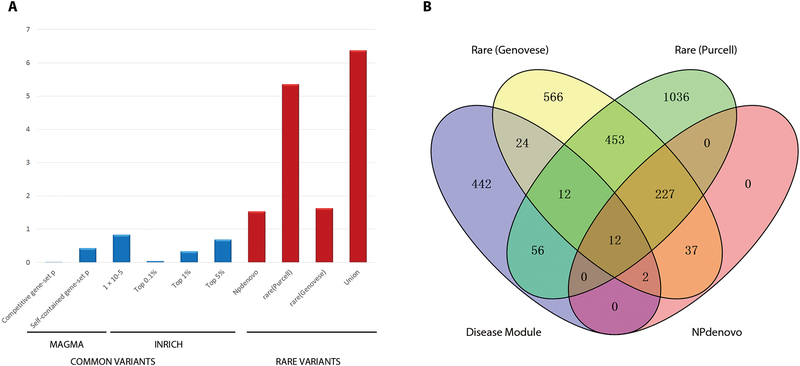
Fig. 2. Enrichment of common and rare genetic variants in the daM for SCZ or BD postmortem brain tissue.
(A) Enrichment of genes in the disease-associated module (daM) with common and rare variants. For common variants, we applied MAGMA and INRICH to test the enrichment. Self-contained gene-set analysis tested whether genes in a gene set showed joint association with SCZ, and competitive gene-set analysis tested whether those genes showed differential association with SCZ compared with other genes in the rest of the genome. Thresholds of 1 × 10−5, top 0.1%, top 1%, and top 5% of all significant SNPs were used as index SNPs in INRICH, and none of them detected significant enrichment. For rare variants, data from two exome sequencing studies and one database were used to test the enrichment. We applied a hypergeometric method to test the overlap with genes collected from Purcell’s study (20), NPdenovo database (19), and the combined gene sets from these two studies. We applied logistic regression to test the rare variant burden using disruptive and damaging ultra-rare variant (dURV) counts for each gene from Genovese’s study (21). (B) The number of genes containing de novo or rare mutations from three sources (19–21) overlapping with genes in daM.