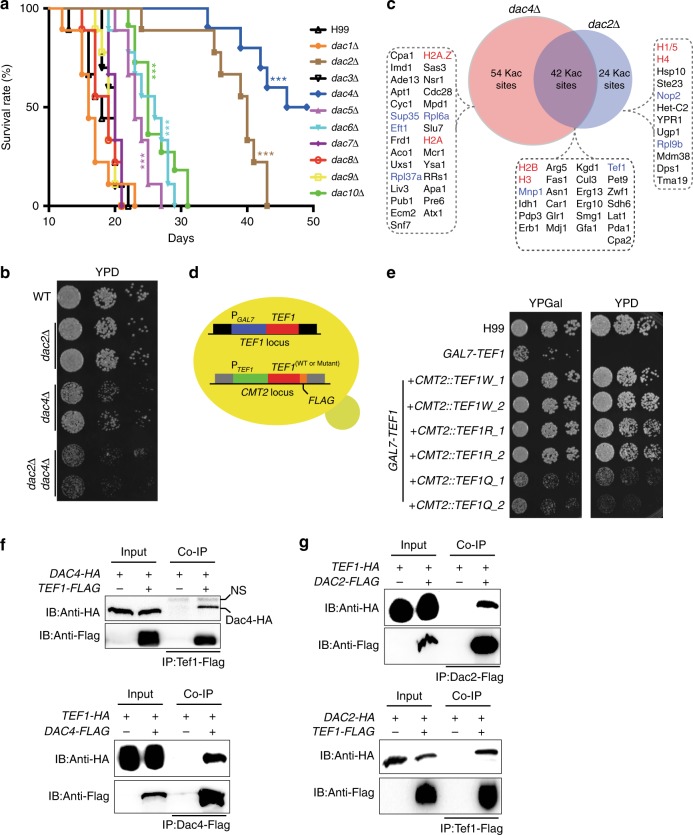
Fig. 3.
C. neoformans deacetylases regulate Tef1 function. a Animal survival assays. Mice (n = 10) were infected with C. neoformans deacetylase mutants. Mice survival rates were recorded. A Kaplan–Meier survival chart was plotted. b Spotting assay of DAC2 and DAC4 mutants. Strains were spotted onto yeast glucose (YPD) agar. The photograph was taken after two days of incubation. c The comparative acetylome analysis of Dac2 and Dac4. Identified proteins were indicated, in which red shows histones and blue indicates proteins involved in protein synthesis. d Generation of C. neoformans TEF1-FLAG strain. The native promoter of TEF1 was replaced with a GAL7 promoter. The resulting strain was transformed with a wild-type TEF1 or a Q or R mutation gene with the native promoter and a C-terminal Flag DNA sequence at the CMT2 locus. e Spotting assays of TEF1 mutants. Strains were spotted onto yeast galactose (YPGal) and YPD agars. The plates were incubated at 30 °C for 2 days. f Co-immunoprecipitation (Co-IP) of Dac4 and Tef1. Co-IP assays were performed in the strains DAC4-HA/TEF1-FLAG and DAC4-FLAG/TEF1-HA. Proteins were pulled down using Flag beads. Anti-HA and anti-Flag antibodies were used. NS indicates nonspecific bands. Full blots are shown in Supplementary Fig. 3e. g Co-IP of Dac2 and Tef1. The assay was performed as described in Fig. 3f, except that the strains DAC2-HA/TEF1-FLAG and DAC2-FLAG/TEF1-HA were used. Full blots are shown in Supplementary Fig. 3e