Abstract
The airways of individuals with cystic fibrosis (CF) are abundantly colonised by Staphylococcus aureus and Pseudomonas aeruginosa. Co-infecting hypoxic regions of static mucus within CF airways, together with decreases in pulmonary function, mucus plugging and oxygen consumption by host neutrophils gives rise to regions of anoxia. This study determined the impact of anaerobiosis upon S. aureus-P. aeruginosa interactions in planktonic co-culture and mixed species biofilms in vitro. Whilst anoxia reduced the ability for P. aeruginosa CF isolates to dominate over S. aureus, this occurred in an isolate dependent manner. Investigations into the underlying mechanisms suggest that the anti-staphylococcal compound facilitating P. aeruginosa dominance under normoxia and anoxia is greater than 3 kDa in size and is heat-stable. Not all interspecies interactions studied were antagonistic, as S. aureus exoproducts were shown to restore and enhance P. aeruginosa motility under normoxia and anoxia in an isolate dependent manner. Collectively, this study suggests changes in oxygen availability within regions of the CF lung is likely to influence interspecies interactions and in turn, potentially influence disease progression.
Subject terms: Microbiology, Bacteriology
Introduction
Cystic fibrosis (CF) is the most common inherited genetic condition in the Caucasian population. An autosomal recessive disorder, CF arises due to mutations in the cystic fibrosis transmembrane conductance regulator (CFTR)1, an ion channel belonging to the ATP-Binding Cassette Transporter family of membrane proteins. Mutations within the CFTR lead to defective apical chloride ion transport, sodium hyperabsorption and a decrease in cell surface liquid2,3. This ultimately results in the production of highly viscous dehydrated mucus, affecting the normal physiology of numerous organs3. Impairment in mucociliary clearance within CF airways predisposes individuals to the development of several opportunistic respiratory infections, typically occurring in a highly-sequential, age-dependent manner4.
A commensal of the anterior nares5, the Gram-positive coccus Staphylococcus aureus (S. aureus) is often the first bacterium to be isolated from the sputum of children with CF and remains one of the most prevalent pathogens throughout the first decade of life6,7. CF airways are colonised by a complex polymicrobial community, with acquisition of the Gram-negative organism Pseudomonas aeruginosa (P. aeruginosa) typically occurring through environmental colonisation8, although patient-patient transmission has been shown to facilitate the spread of drug resistant epidemic strains9,10. As an individual progresses through adolescence and into early adulthood, P. aeruginosa replaces S. aureus as the dominant pathogen in CF airways7,11. There is temporal phenotypic diversity of P. aeruginosa during the course of CF airway infection12,13. Compared to the early infecting environmental strain, chronic infection with P. aeruginosa is associated with a loss of motility, loss of quorum sensing, the acquisition of the mucoid phenotype and increased resistance to antibiotics14,15.
The ability of P. aeruginosa to dominate has been attributed to an arsenal of extracellular virulence properties, several of which target S. aureus. The staphylolytic protease elastase A (LasA) degrades the peptidoglycan cell wall of S. aureus16, whilst the phenazine pyocyanin and secondary metabolites hydrogen cyanide and 2-heptyl-4-hydroxyquinoline N-oxide (HQNO) inhibit S. aureus oxidative respiration, consequently slowing its growth17–19. Despite this antagonism, S. aureus is detected in a third of CF adults who are culture positive for P. aeruginosa7,20, with two recent studies suggesting S. aureus and P. aeruginosa occupy identical regional niches of the CF lung21,22. Co-infection of CF airways with S. aureus and P. aeruginosa has been associated with decreased pulmonary function and an increased frequency of pulmonary exacerbations compared to mono-infection20,23.
Studies assessing polymicrobial CF airway infection to date, including S. aureus-P. aeruginosa interactions, have been conducted under normoxia (21% environmental oxygen), with vigorous culture aeration (200–250 rpm)18,24–29. However, increases in CF epithelia respiration, thick mucus plugging, decreased pulmonary function and oxygen consumption by polymicrobial communities and host neutrophils encourage the development of a static growth environment, with very low to absent levels of oxygen30–34. The presence of obligate anaerobes has also been shown to contribute to disease severity and inflammation in CF airways35–37.
Though P. aeruginosa is a facultative anaerobe that preferably uses aerobic respiration, its ability to undergo anaerobiosis is due to the presence of nitrate (NO3−), which acts as an end terminal electron acceptor38,39. Nitrate has been detected in both airway surface liquid (ASL) and in CF airway sputum31,32,40. A microarray study of P. aeruginosa obtained from CF sputum detected genes essential for P. aeruginosa denitrification41, whilst sera from individuals with CF have also been shown to contain antibodies to P. aeruginosa proteins involved in denitrification42.
Whilst anaerobiosis has previously been shown to influence the P. aeruginosa phenotype including growth, virulence and sensitivity to antibiotics31,43–48, its impact upon the interactions of P. aeruginosa with other CF pathogens has yet to be investigated. Initial experiments in this study sought to determine the impact of normoxia and anoxia upon the in vitro interactions between S. aureus and CF clinical isolates of P. aeruginosa in planktonic co-culture and mixed species biofilms, the two modes of P. aeruginosa growth known to occur in the CF lung. To provide insight into the mechanisms driving interspecies interactions under different oxygen conditions, the impact of anaerobiosis upon the production of several anti-staphylococcal exoproducts produced by P. aeruginosa were studied, which was further assessed using size fractionation and heat-treatment. Lastly, the impact of anaerobiosis and S. aureus exoproducts upon P. aeruginosa motility was determined. Data from this study suggest that oxygen availability exerts an isolate dependent effect upon S. aureus-P. aeruginosa interactions that influences the predominance of each organism within different regions of the CF lung.
Results
Effect of oxygen availability upon S. aureus-P. aeruginosa competition in planktonic co-culture
We sought to identify whether changes in oxygen availability influenced S. aureus-P. aeruginosa interactions in planktonic (free-swimming) culture. Reference strain P. aeruginosa PAO1 and P. aeruginosa CF isolates 5, 6 and 7 were selected for analysis following the growth inhibition results of the agar plate cross-streak assay (shown in Supplementary Fig. S1). PAO1 and P. aeruginosa CF clinical isolates 5 and 7 were able to inhibit S. aureus under normoxia, whilst CF isolate 6 had no effect upon the growth of S. aureus. Additionally, preliminary experiments demonstrated that these CF isolates differed with regards to protease production (assessed on skimmed milk agar) and pyocyanin (following bacterial culture upon Pseudomonas isolation agar). S. aureus ATCC 6538 was chosen due to it being an accessible and widely used laboratory reference strain. In addition to being known to produce biofilm, (important for the co-species biofilm studies)49, it has also been previously used to study S. aureus-P. aeruginosa interactions50. Furthermore, it has had its complete genome sequenced51, which may aid in determining the virulence properties it is likely to exhibit.
Density matched S. aureus and P. aeruginosa cultures were inoculated at an equal ratio, with planktonic growth competition being assessed over 24 h, comparing the colony counts of the bacteria grown in pure culture, to those grown in co-culture. To clearly compare the differences of growth in monoculture with co-culture, the competitive index (CI) and relative increase ratio (RIR) were calculated.
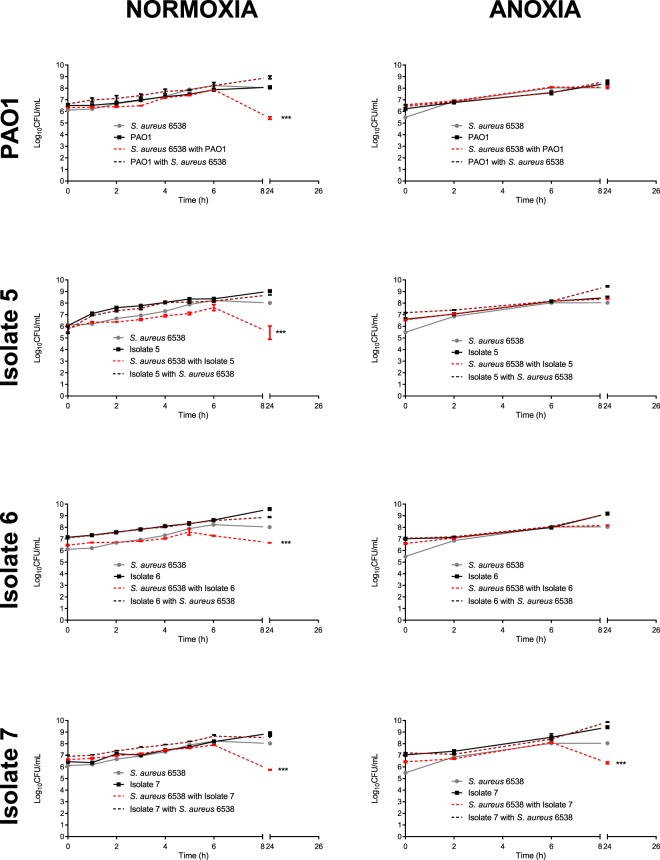
As shown in Fig. 1 (left panel), all P. aeruginosa isolates tested were able to outcompete S. aureus under normoxia. PAO1 and CF isolates 5 and 7 caused an approximate 2.5 log reduction in viable S. aureus at 24 h (P < 0.001), whilst CF isolate 6 caused an approximate 1.5 log reduction in S. aureus at 24 h (P < 0.001). The CI of PAO1 versus S. aureus was significantly different from the RIR at 2, 3, 5, 6 and 24 h, whilst the CI for all CF isolates was significantly higher than the RIR at 6 and 24 h time points only (Supplementary Fig. S2). S. aureus killing was incomplete under normoxia for PAO1 and the CF isolates tested, with S. aureus still being detected at high bacterial counts at 24 h.
Figure 1.
S. aureus-P. aeruginosa competition in planktonic co-culture under normoxia and anoxia. S. aureus and P. aeruginosa were grown statically at 37 °C for 24 h in either single or dual culture, under normoxia or anoxia. At regular intervals, aliquots were taken and plated onto selective agar. Each data point represents the mean ± S.E.M of three independent experiments (N = 3), each performed in triplicate. Statistical differences were determined using a two-way ANOVA with Bonferroni post-hoc, comparing the CI to the RIR at each time point ***P < 0.001.
Similar experiments were conducted to determine the effects of anoxia upon co-culture competition (Fig. 1, right panel). S. aureus growth was unaffected by the presence of P. aeruginosa PAO1 and CF isolates 5 and 6 at all time points tested (see Supplementary Fig. S2), whilst CF isolate 7 retained its ability to dominate, causing an approximate 1 log reduction in the growth of S. aureus at 24 h compared to monoculture (P < 0.001).
Oxygen availability and community composition in mixed species biofilms
As S. aureus and P. aeruginosa often co-exist within a polymicrobial biofilm within the CF lung, we determined the impact of oxygen availability upon the number of viable bacteria within single and mixed biofilms.
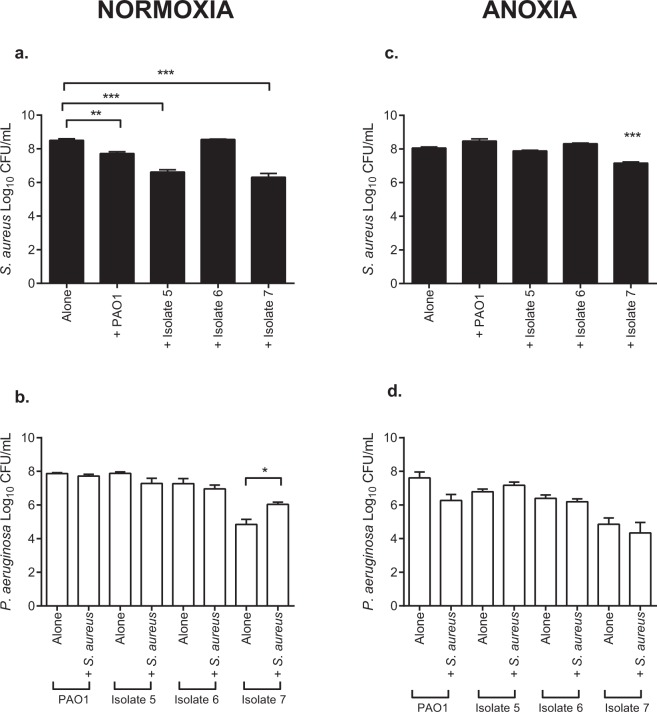
Under normoxia (Fig. 2a), S. aureus numbers were reduced in the presence of PAO1 (P < 0.01) and CF isolates 5 and 7 (P < 0.001), compared to S. aureus alone. Unlike in planktonic culture (Fig. 1), CF isolate 6 was unable to outcompete S. aureus in mixed species biofilm. Furthermore, S. aureus was unable to exert an antagonistic affect upon any of the P. aeruginosa isolates tested (Fig. 2b), with isolates of P. aeruginosa being recovered in numbers comparable to those isolated in single species biofilm. CF isolate 7 however, demonstrated a significant increase in CFU/mL in mixed species biofilm compared to single culture (P < 0.05).
Figure 2.
S. aureus and P. aeruginosa sessile viability in mono- and co-culture biofilms under normoxia and anoxia. Bacteria were grown in 96-well flat bottom plates either individually or in a 1:1 ratio for 24 h at 37 °C under static normoxia or anoxia. Biofilms were washed, detached, vortexed, serially diluted and plated onto selective agar for quantification. Bars represent the mean ± S.E.M of three individual experiments (N = 3), each performed in triplicate. Statistical differences were determined using one-way ANOVA with Tukey’s post-hoc. *P < 0.05, **P < 0.01, ***P < 0.001.
Following growth under anoxia, PAO1 and CF isolates 5 and 6 were no longer able to outcompete S. aureus in mixed species biofilm (Fig. 2c) whilst CF isolate 7 retained its ability to reduce S. aureus viability (P < 0.001). All CF isolates tested were unaffected by the presence of S. aureus under anoxia with P. aeruginosa being enumerated at numbers similar to those in P. aeruginosa monoculture biofilms (Fig. 2d). The data for S. aureus-P. aeruginosa competition in planktonic co-culture and mixed species biofilms is summarised in a table in Supplementary Table S1.
Impact of oxygen upon anti-staphylococcal exoproducts
It is evident that anoxia facilitates S. aureus-P. aeruginosa co-existence in an isolate dependent manner, in both planktonic co-culture and mixed species biofilms. To determine how oxygen availability permits bacterial co-existence and why P. aeruginosa CF isolate 7 was still able to reduce S. aureus viability, the production of several known anti-staphylococcal exoproducts in cell-free supernatants of P. aeruginosa were assessed following growth under normoxia and anoxia. This study focused upon the production of proteases, the lysis of S. aureus, the production of the respiratory inhibitor pyocyanin, the siderophore pyoverdine and biosurfactants.
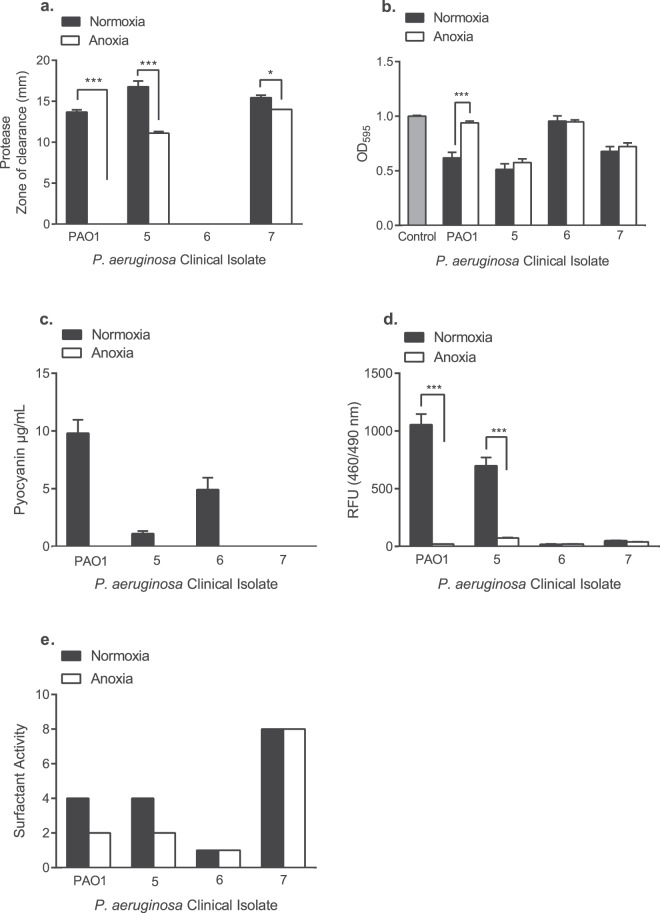
As P. aeruginosa is known to secrete a number of proteases, skimmed milk agar has previously been used to study the total protease activity of P. aeruginosa50,52. The diameter of the zones of clearance were measured in millimetres (mm). As shown in Fig. 3a, PAO1 and CF isolates 5 and 7 produced detectable levels of protease under normoxia. Following growth under anoxia, CF isolates 5 and 7 retained their protease activity, whilst this was below the limit of detection for PAO1. CF isolate 6 failed to produce detectable proteases following growth under normoxia and anoxia.
Figure 3.
Impact of normoxia and anoxia upon the production of anti-staphylococcal exoproducts by P. aeruginosa. (a) Protease production was determined following the addition of P. aeruginosa cell-free supernatants to wells in milk agar, prior to the diameter of the zones of clearance (mm) being measured following 24 h incubation. (b) P. aeruginosa cell-free supernatants were added to heat-killed S. aureus and staphylolytic activity was determined by measuring changes in the OD595 after 60 min. The control consisted of heat-killed S. aureus with sterile LBN broth only. (c) Pyocyanin was extracted from P. aeruginosa cell-free supernatants using chloroform and 0.2 M hydrochloric acid. Spectrophotometric measurements were taken at OD520 and multiplied by extinction co-efficient of pyocyanin at this wavelength (17.072) to express pyocyanin in µg/mL. (d) Pyoverdine production was quantified by measuring the relative fluorescence units (RFU) of P. aeruginosa cell-free supernatants following excitation at 460 nm and emission at 490 nm. (e) Surfactant activity was measured using the established drop collapse assay, where cell-free supernatants were serially diluted (1:1) with deionised water containing 0.005% crystal violet to aid visualisation. Surfactant scores are equal to the reciprocal of the greatest dilution at which there was surfactant activity. Data shown are the mean ± S.E.M. of three independent experiments (N = 3), each performed in triplicate. In panels A, B and D, statistical differences were determined using one-way ANOVA with Tukey’s post-hoc test *P < 0.05, **P < 0.01 and ***P < 0.001.
P. aeruginosa PAO1 has previously been shown to lyse S. aureus16. Thus, the ability of P. aeruginosa CF isolates to lyse heat-killed S. aureus was also determined following growth under normoxia and anoxia, a method used previously53–55. As shown in Fig. 3b, CF isolate 6 was the only isolate unable to significantly lyse heat-killed S. aureus under normoxia (P > 0.05). Following growth under anoxia (Fig. 3b), PAO1 lost its ability to significantly lyse heat-killed S. aureus compared to normoxia (P < 0.001), whilst CF isolates 5 and 7 retained their significant staphylolytic activity under anoxia. CF isolate 6 did not lyse heat-killed S. aureus under anoxia.
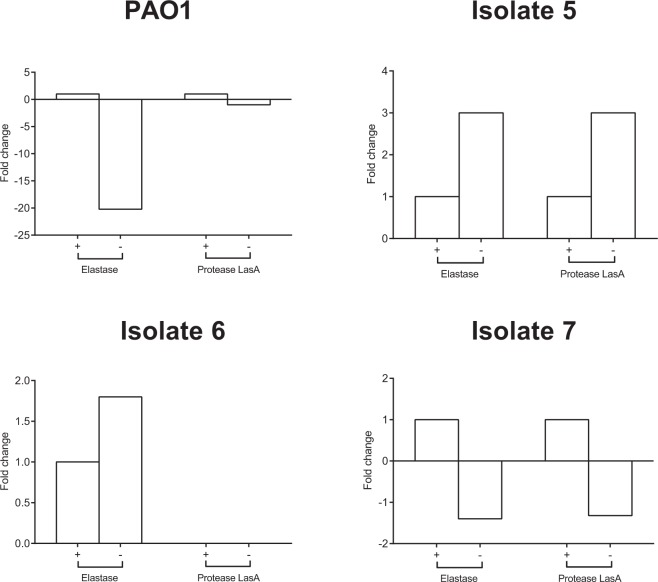
The complete cell-free secretomes of P. aeruginosa PAO1 and CF isolates 5, 6 and 7 are shown in Supplementary Tables S2 to S5. Expression levels of elastase and protease LasA following growth under normoxia and anoxia were studied by mass spectrometry (Fig. 4) to determine whether there was any correlation with the protease and staphylolytic activity shown in Fig. 3a,b. As shown in Fig. 4, whilst all of the isolates tested secreted detectable levels of elastase under both environmental conditions, CF isolate 6 was the only isolate which did not produce detectable levels of protease LasA under both normoxia and anoxia. Analysis of the PAO1 cell-free secretome demonstrated a 20.2-fold decrease in the relative abundance of elastase under anoxia, whilst exhibiting a minimal fold change in protease LasA. Conversely, CF isolates 5 and 7 exhibited a 3.0-fold increase and 1.4-fold decrease in elastase respectively, along with a 3.0-fold increase and 1.3-fold decrease in protease LasA. Once more, CF isolate 6 failed to exhibit staphylolytic activity under anaerobiosis (Fig. 4b), with no protease LasA being detected in its secretome (Fig. 4).
Figure 4.
Detection of elastase and protease LasA in the P. aeruginosa cell-free secretome. Cell-free supernatants obtained from 24 h cultures of PAO1 and CF isolates 5, 6 and 7 grown under normoxia (+) and anoxia (−) were subject to protein precipitation, SDS-PAGE electrophoresis and MS analysis. Data represents N = 1 from five pooled samples. The abundance of elastase and protease LasA is expressed as fold-change compared to normoxia.
The production of the respiratory inhibitor pyocyanin was also determined. Under normoxia, PAO1 and CF isolates 5 and 6 produced varying degrees of pyocyanin following phenol-chloroform extraction, whilst this was below the limit of detection for CF isolate 7 (Fig. 3c). No pyocyanin was detected for any of the P. aeruginosa isolates following growth under anoxia.
As shown in Fig. 3d, P. aeruginosa PAO1 and CF isolate 5 produced pyoverdine under normoxia, with this being significantly reduced under anoxia (P < 0.001). CF isolates 6 and 7 produced minimal levels of pyoverdine, which was unaffected by anoxia. Lastly, PAO1 and all CF isolates tested exhibited varying degrees of biosurfactant activity, with this being the greatest for CF isolate 7 (Fig. 3e). Anoxia reduced the surfactant activity of PAO1 and CF isolate 5, but not for CF isolates 6 and 7. The data shown in Fig. 3 demonstrates that each P. aeruginosa CF isolate produces a varying array of anti-staphylococcal exoproducts under normoxia, with their production being further influenced by anoxia.
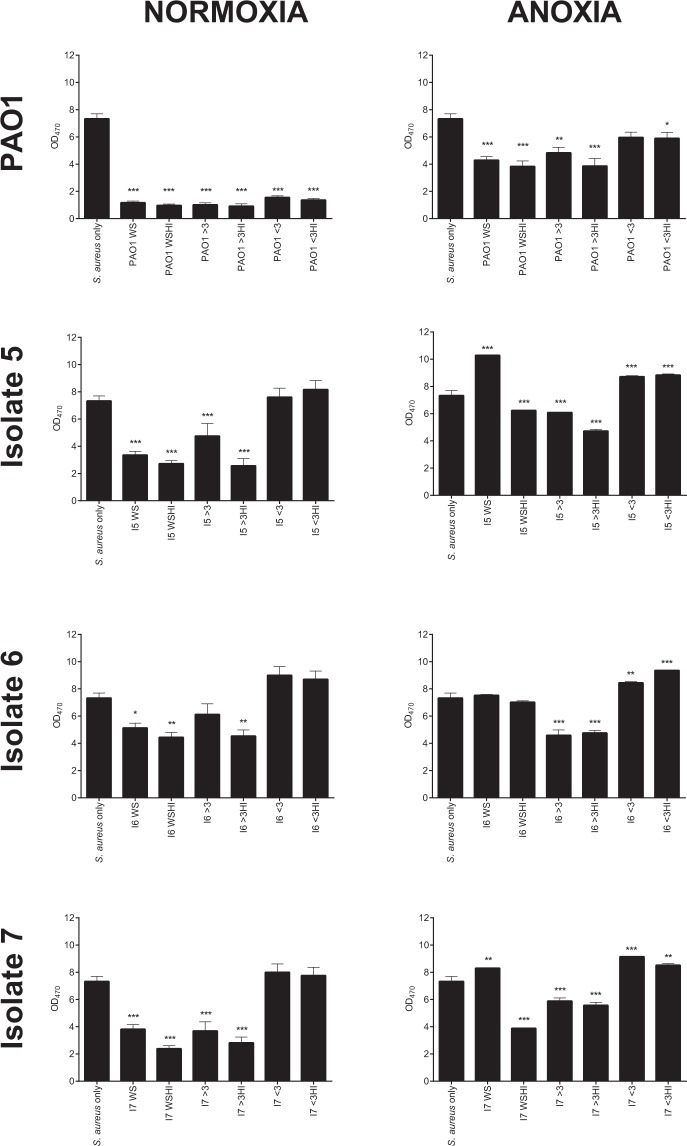
Using these data, we sought to greater determine the nature of the P. aeruginosa anti-staphylococcal compound which mediates P. aeruginosa dominance under both normoxia and anoxia. Cell-free supernatants from P. aeruginosa cultures were passed through 3 kDa molecular weight cut-off filters, in an effort to determine the size of the anti-staphylococcal compound. Whilst the 20 kDa protease LasA would be retained within the >3 kDa fraction, smaller virulence factors such as pyocyanin, rhamnolipids and pyoverdine would pass into the <3 kDa fraction. Heat-treatment was used to determine whether the anti-staphylococcal compound was proteinaceous in nature, as boiling would be expected to abolish its anti-staphylococcal activity. Cell-free culture supernatants were subsequently added to log-phase cultures of S. aureus to assess their ability to inhibit S. aureus planktonic growth over 14 h.
As shown in Fig. 5, under normoxia, P. aeruginosa PAO1 whole supernatant, >3 kDa and <3 kDa fractions were able to significantly reduce S. aureus growth compared to S. aureus alone (P < 0.001). This anti-staphylococcal activity was not lost following heat-treatment of the fractions. Interestingly, S. aureus antagonism across all the CF clinical isolates tested under normoxia were restricted to the whole supernatant and >3 kDa fractions only, which was unaffected by heat-treatment.
Figure 5.
Heat-treatment and size fractionation of P. aeruginosa cell-free supernatants upon the growth of S. aureus. Cell-free supernatants from P. aeruginosa grown under normoxia or anoxia were added to standardised S. aureus directly or were first subjected to 3 kDa size fractionation, with select fractions then being subjected to boiling. Plates were incubated at 37 °C for 14 h and the OD470 of S. aureus cultures read at hourly intervals. Plots represent the mean OD at 14 h ± S.D. of two individual experiments (N = 2), each performed in duplicate. WS: Whole supernatant, WSHI: Whole supernatant heat-inactivated, >3: greater than 3 kDa, >3HI: greater than 3 kDa heat-inactivated, <3: less than 3 kDa, <3HI: less than 3 kDa heat-inactivated. Statistical differences were determined using one-way ANOVA with Dunnett’s post-hoc (versus S. aureus only control). *P < 0.05, **P < 0.01 ***P < 0.001.
Under anoxia (Fig. 5) significant anti-staphylococcal activity was detected in all but the untreated <3 kDa fraction of PAO1. Once again, the anti-staphylococcal activity of the fractions was unaffected by heat-treatment. The heat-treated whole culture supernatant and both the untreated and heat-treated >3 kDa fractions obtained from CF isolate 5 significantly reduced S. aureus growth. For CF isolate 6, only the >3 kDa fractions significantly reduced S. aureus growth, where once again, heat-treatment failed to abolish this activity. As seen with CF isolate 5, the heat-treated whole culture supernatant and the untreated and heat-treated >3 kDa fractions obtained from CF isolate 7 were the most inhibitory towards S. aureus growth. Collectively, this suggests that the major anti-staphylococcal exoproduct produced by P. aeruginosa is heat-stable and >3 kDa in size.
Proteomic analysis of the cell-free secretomes of PAO1 and CF isolates 5, 6 and 7 (Supplementary Tables S2 to S5) did not identify any obvious lead candidate for the active factor which is likely to mediate P. aeruginosa dominance under normoxia and anoxia. Whilst the mass spectrometry experiments performed were used to generate a proteomic dataset only, it is possible that the extracellular anti-staphylococcal factor is not proteinaceous in nature. Thus, further work is required in order to identify the active anti-staphylococcal factor.
Impact of S. aureus exoproducts and anoxia upon P. aeruginosa motility
Lastly, as S. aureus infection precedes P. aeruginosa, we sought to determine the influence of S. aureus exoproducts upon P. aeruginosa swimming and swarming motilities. Both free and surface attached forms of motility are likely to play important roles in airway colonisation and disease progression. The impact of anoxia upon P. aeruginosa motility in the presence and absence of S. aureus exoproducts was also determined.
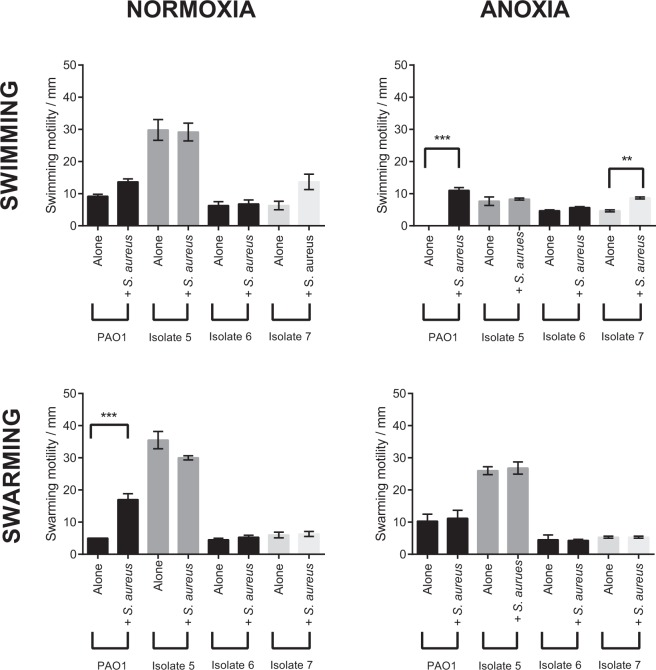
PAO1 and the CF isolates tested exhibited varying degrees of swimming motility under normoxia (Fig. 6). CF isolate 5 alone exhibited significantly greater swimming motility under normoxia compared to PAO1, CF isolate 6 and CF isolate 7 alone (P < 0.001). P. aeruginosa motility was unaffected by the inclusion of S. aureus exoproducts. Under anoxia, CF isolate 5 alone exhibited the greatest swimming motility compared to PAO1 alone (P < 0.01) and CF isolates 6 and 7 alone (P < 0.05). Compared to normoxia, anoxia reduced the swimming motility of PAO1 alone (P < 0.01), CF isolate 5 alone (P < 0.001) and CF isolate 5 in the presence of S. aureus (P < 0.001). PAO1 alone lost its swimming motility under anoxia whilst the addition of S. aureus cell-free supernatant significantly restored the swimming motility of PAO1 (P < 0.001). S. aureus cell-free supernatant also enhanced the swimming motility of CF isolate 7 compared to CF isolate 7 alone (P < 0.01).
Figure 6.
Effect of S. aureus culture supernatant and oxygen upon P. aeruginosa motility. Swimming and swarming motilities of P. aeruginosa isolates were assessed following an overnight incubation on swimming and swarming agar plates. To test the effects of S. aureus culture supernatant upon these two forms of motility, a 1:100 dilution of the culture supernatant was added to plates before the agar set. Data are presented as mean ± S.E.M from three independent experiments each performed in duplicate. Statistical differences were determined using one-way ANOVA with Tukey’s post-hoc. **P < 0.01 ***P < 0.001.
Under normoxia, CF isolate 5 alone exhibited the greatest swarming motility compared to PAO1 and CF isolates 6 and 7 alone (P < 0.001) (Fig. 6). The addition of S. aureus exoproducts only enhanced the swarming motility of PAO1 (P < 0.001). Under anoxia, CF isolate 5 exhibited the greatest swarming motility compared to PAO1, including CF isolates 6 and 7 alone (P < 0.001). S. aureus exoproducts did not modulate P. aeruginosa swarming motility under anoxia. Only CF isolate 5 alone exhibited a significant reduction in swarming motility under anoxia compared to normoxia (P < 0.01).
As S. aureus exoproducts were shown to modulate the swimming motility of PAO1 and CF isolate 7 under anoxia, including the swarming motility of PAO1 under normoxia (Fig. 6), S. aureus cell-free supernatants were subjected to size fractionation and heat-treatment to determine the size and nature of the effector compound(s). As shown in Fig. 7, all fractions regardless of size and heat-treatment were able to modulate P. aeruginosa motility to the same extent as the whole culture supernatants (Fig. 6).
Figure 7.
Heat-treatment and size fractionation of S. aureus cell-free supernatants upon P. aeruginosa swimming and swarming motility. Swimming and swarming motilities of the select P. aeruginosa isolates were assessed following an overnight incubation on swimming and swarming agar plates. S. aureus only motility values were taken from Fig. 6. WS: whole supernatant fractions of S. aureus culture supernatant were taken from Fig. 6. Plates were supplemented with a 1:100 dilution of S. aureus culture supernatant fractions (>3 or <3 kDa), with select fractions also being subject to heat-treatment. >3: greater than 3 kDa, >3HI: greater than 3 kDa heat-inactivated, <3:less than 3 kDa, <3HI: less than 3 kDa heat-inactivated. Data are presented as mean ± S.D from two independent experiments (N = 2) each performed in duplicate. Statistical differences were determined using one-way ANOVA with Dunnett’s post-hoc (versus WS: whole supernatant only).
Discussion
It is now appreciated that mucus plugging, decreases in pulmonary function and the consumption of oxygen by polymicrobial communities and host neutrophils create microenvironments in CF airways which are devoid of oxygen31,56. Whilst previous studies have demonstrated anaerobiosis attenuates the growth and virulence of P. aeruginosa laboratory strains and clinical isolates43,44, this study aimed to provide novel insights into the impact of anaerobiosis upon the interactions of S. aureus and CF clinical isolates of P. aeruginosa in planktonic co-culture and mixed species biofilms, including the possible mechanisms governing interspecies interactions under both environmental conditions.
Our competition data showed oxygen availability exerts a major role in S. aureus and P. aeruginosa competition in both planktonic co-culture and mixed species biofilms. Under normoxia (Fig. 1), PAO1 and all CF isolates 5, 6 and 7 were all able to outcompete S. aureus in planktonic co-culture at 24 h, without their own growth being adversely affected. This finding is supported by other in vitro studies conducted under normoxia where P. aeruginosa CF isolates were shown to reduce the viability of S. aureus in planktonic culture25,27,57. In all planktonic 24 h co-culture experiments, P. aeruginosa was unable to completely kill S. aureus.
PAO1 and CF isolates 5 and 7 retained their ability to antagonise S. aureus growth in mixed species biofilms under normoxia, whilst CF isolate 6 did not (Fig. 2). The inability of CF isolate 6 to outcompete S. aureus in mixed species biofilms as in planktonic co-culture (Figs 1 and 2) is potentially explained by previous studies which demonstrate differences in bacterial virulence following growth in planktonic and biofilm culture58–60.
The introduction of anoxia caused PAO1 and CF isolates 5 and 6 to lose their ability to outcompete S. aureus in both planktonic co-culture and mixed species biofilms (Figs 1 and 2), where S. aureus was enumerated in densities seen in monoculture. These results support those of a previous study which demonstrated anaerobiosis reduced the ability of P. aeruginosa PAO1 to induce zones of clearance upon confluent bacterial lawns of Staphylococcus epidermidis61. Furthermore, the culture of P. aeruginosa strains under hypoxia (1% oxygen) has previously been shown to attenuate P. aeruginosa virulence in an in vivo model of infection compared to normoxia through decreased expression of several virulence factors62. From these data, it is possible that anoxia is one of several mechanisms facilitating isolate specific S. aureus-P. aeruginosa co-existence in the CF lung.
Interestingly, CF isolate 7 was the only P. aeruginosa isolate able to reduce S. aureus viability under anoxia, in both planktonic co-culture and mixed species biofilm (Figs 1 and 2). To greater understand this phenomenon, the production of several anti-staphylococcal exoproducts was assessed across isolates, following growth under normoxia and anoxia. The use of molecular weight cut off filters and heat-treatment of the cell-free fractions served to determine the size and nature of the anti-staphylococcal compound likely to be mediate P. aeruginosa dominance under normoxia and anoxia.
PAO1 has previously been shown to produce LasA, responsible for cleaving the peptidoglycan cell wall of S. aureus24,63. Unlike CF isolate 6, PAO1 and CF isolates 5 and 7 were able to lyse heat-killed S. aureus under normoxia (Fig. 3b). PAO1 lost its staphylolytic ability under anoxia however (Fig. 3b), a finding supported by reports of a decrease in the transcription of LasA for PAO1 under anoxia64 and a reduction in PAO1 elastase B (LasB) production under anoxia44. Perhaps the LasA produced by PAO1 following growth under anoxia is below a threshold to exert a considerable affect upon S. aureus viability compared to that produced by CF isolates 5 and 7, although further work is required to determine this. S. aureus has also previously been shown to exhibit an increase in cell wall thickness under anaerobiosis65 and whilst this was not investigated in this study, this mechanism may further reduce S. aureus susceptibility to P. aeruginosa LasA under anoxia.
Whilst LasA may be advantageous to P. aeruginosa under growth in polymicrobial communities with S. aureus, collectively, LasA mediated lysis of S. aureus alone does not appear to be essential for facilitating P. aeruginosa dominance under normoxia. All CF isolates were able to outcompete S. aureus in planktonic co-culture (Fig. 1), yet CF clinical isolate 6 failed to exhibit detectable staphylolytic activity (Fig. 3b) or secrete detectable protease LasA as determined by mass spectrometry (Fig. 4). LasA also does not appear to be essential for modulating P. aeruginosa dominance under anoxia, as CF isolates 5 and 7 both retained their staphylolytic activity under anoxia (Fig. 3b). Only CF isolate 7 was able to outcompete in planktonic co-culture (Fig. 1) and mixed species biofilm (Fig. 2). Furthermore, whilst whole culture supernatants and >3 kDa fractions from PAO1 and the CF isolates were able to antagonise S. aureus growth, heat-treatment failed to abolish their inhibitory activity (Fig. 5). As staphylolysis is known to be mediated by LasA, boiling such fractions would be expected to denature this 20 kDa protease.
The role of LasA in governing interspecies interactions may also be complicated by previous findings which demonstrate how the growth of S. aureus in the presence of P. aeruginosa LasA and staphylolysin produced by Staphylococcus staphylolyticus, selects for S. aureus colonies which lack a cell wall66,67. Whilst the emergence of such colonies in CF airways requires further study, selection of such a phenotype may serve to facilitate S. aureus persistence under both normoxia and anoxia in the presence of P. aeruginosa.
As previously mentioned, pyocyanin inhibits S. aureus oxidative respiration, causing a reduction in growth. PAO1 and CF isolates 5 and 6 produced detectable levels of pyocyanin under normoxia following phenol-chloroform extraction, whilst this was below the limit of detection for CF isolate 7 (Fig. 3c). Despite this, CF isolate 7 was still able to dominate S. aureus under normoxia in planktonic co-culture (Fig. 1) and mixed species biofilms (Fig. 2). Furthermore, pyocyanin was below the limit of detection for all culture supernatants cultured under anoxia following phenol-chloroform extraction (Fig. 3c), yet CF isolate 7 was still able to dominate. Whilst the inability to detect pyocyanin under anoxia may be due to the redox nature of the phenazine68, a previous study demonstrated that hypoxia significantly reduced pyocyanin production by P. aeruginosa69.
Proteins involved in phenazine biosynthesis were only detected in the secretomes of P. aeruginosa PAO1 and P. aeruginosa CF isolate 5. As shown in Supplementary Table S2, P. aeruginosa PAO1 exhibited a 10.4-fold decrease in phenazine biosynthesis protein PhzB 1 and a 12.3-fold decrease in phenazine biosynthesis protein PhzB 2 respectively under anoxia. Additionally, there was a 21.0-fold decrease in Phenazine biosynthesis protein PhzB 1 for P. aeruginosa CF isolate 5 under anoxia (Supplementary Table S3). These data support the phenotypic data relating to pyocyanin production shown in Fig. 3c.
Size exclusion and heat-treatment of the P. aeruginosa cell-free supernatants also demonstrated that S. aureus antagonism was restricted to the >3 kDa fraction and was not abolished following heat-treatment (Fig. 5). However, it is expected that pyocyanin would be present in the <3 kDa fraction and that heat-treatment would abolish its activity.
Pyoverdine production by P. aeruginosa is known to reduce S. aureus growth, and enhance P. aeruginosa virulence27,29,70. As shown in Fig. 3d, PAO1 and CF isolate 5 produced large amounts of pyoverdine. Furthermore, anoxia was shown to significantly reduce pyoverdine production by PAO1 and CF isolate 5 compared to normoxia, a finding also reported following the growth of P. aeruginosa under hypoxia69. Despite CF isolates 6 and 7 producing minimal amounts of pyoverdine under normoxia (Fig. 3d), all P. aeruginosa CF isolates were able to outcompete S. aureus in planktonic co-culture (Fig. 1). Additionally, CF isolate 7 was able to outcompete S. aureus under anoxia despite producing minimal amounts of pyoverdine. As the antimicrobial activity of P. aeruginosa cell-free supernatants was restricted to the >3 kDa fractions (Fig. 5), it is unlikely that siderophore production in isolation drives S. aureus antagonism. However, we recognise the importance of pyoverdine in governing interspecies interactions may be underestimated in this study as iron-depleted growth media was not used to culture P. aeruginosa.
The surfactant activity of P. aeruginosa rhamnolipids has been previously shown to target S. aureus and reduce its viability71–73. Our data demonstrated that PAO1 and the CF P. aeruginosa isolates tested harboured biosurfactant activity under normoxia, with this being the greatest for CF isolate 7 (Fig. 3e). Whilst CF isolate 7 retained its high surfactant score under anoxia, size exclusion studies demonstrated that the anti-staphylococcal activity for all isolates was restricted to the >3 kDa fraction under both normoxia and anoxia (Fig. 5) and yet it is expected that the rhamnolipids would be retained within the <3 kDa fractions.
Collectively, it is reasonable to suggest from the results of this study that the anti-staphylococcal compound which facilitates P. aeruginosa dominance under both normoxia and anoxia is heat-stable, >3 kDa in size and is likely not one of the anti-staphylococcal factors studied. An interesting observation in Fig. 5 is that the heat-treated whole supernatant and >3 kDa fractions for most P. aeruginosa CF isolates under normoxia and anoxia appeared to more greatly reduce S. aureus growth compared to those which had not been heat treated. It is reasonable that heating may either increase the activity of the anti-staphylococcal factor, or alternatively lead to the loss of a factor within the supernatant which may otherwise protect S. aureus directly or indirectly. We also recognise that a limitation of this study is that the absolute concentration of the P. aeruginosa exoproducts in cell-free culture supernatants was not determined and it may be that some of the virulence factors were at concentrations too low to exert an antagonistic effect upon S. aureus.
P. aeruginosa is known to employ several other virulence properties which were not characterised in this study and may contribute to the differences seen in bacterial competition. This includes the respiratory inhibitor hydrogen cyanide74. However, its role in governing S. aureus-P. aeruginosa interactions is likely to be minimal in this study as it would be found in the <3 kDa fraction, and yet these fractions did not exert considerable anti-staphylococcal activity upon exposure to planktonic S. aureus under normoxia (Fig. 5). Furthermore, previous research has demonstrated that hydrogen cyanide is rapidly inactivated under strict anoxia75 and thus its ability to reduce S. aureus viability under anoxia is considered minimal.
P. aeruginosa also produces the respiratory inhibitor HQNO, which is known to reduce S. aureus viability19,27,76. Its production is regulated by Pseudomonas quinolone signal (PQS)77. PQS production is known to be suppressed under anoxia61,78. As shown in Supplementary Table S5, PQS proteins were only detected in the secretome of P. aeruginosa CF isolate 7. Whilst there was a 9.4-fold decrease in PqsB under anoxia, there was a 4.5-fold increase in PqsC under anoxia. Thus, further work is required to determine the role of HQNO in governing S. aureus-P. aeruginosa interspecies interactions.
It is also reasonable to suggest that the ability of P. aeruginosa to dominate under both normoxia and anoxia is not due to the secretion of a single anti-staphylococcal factor, but is due to the production of many20,27,79–81. A previous study has reported that deletion of a single virulence property has been shown to reduce P. aeruginosa antagonism towards S. aureus82. The production of alginate by P. aeruginosa has been reported to facilitate S. aureus-P. aeruginosa co-existence within the CF lung, with the overproduction of alginate inhibiting the synthesis secretion of pyoverdine, HQNO and rhamnolipids82. However, as none of the P. aeruginosa CF isolates used in this study were mucoid, alginate is unlikely to play a role in facilitating interspecies co-existence.
Finally, we also show how not all S. aureus-P. aeruginosa interactions studied are antagonistic. Whilst S. aureus has previously been shown to enhance P. aeruginosa antibiotic resistance83,84, we have demonstrated that S. aureus exoproducts were able to restore and enhance P. aeruginosa swimming and swarming motility in an isolate dependent manner under normoxia and anoxia (Fig. 6).
Previous research has demonstrated that the intracellular accumulation of the signalling molecule bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) favours P. aeruginosa biofilm formation85 through the production of exopolysaccharide, whilst repressing flagella motility. Conversely, intracellular decreases in c-di-GMP inhibits exopolysaccharide production and promotes flagella mediated motility86,87. These effects of intracellular c-di-GMP are mediated through its ability to bind and modulate the transcriptional regulator FleQ88,89. It is possible that one or more S. aureus exoproducts within the cell-free supernatant may lower the intracellular concentration c-di-GMP in P. aeruginosa, promoting motility (Fig. 6). Additional work is required as the modulating ability of S. aureus culture supernatant upon P. aeruginosa motility was not seen in all isolates and could not be restricted to a single fraction (Fig. 7).
Twitching motility in the presence and absence of S. aureus exoproducts was also investigated but in our hands no twitching motility was detected in any strains. As anoxia was also shown to influence P. aeruginosa motility, the presence of S. aureus and changes in oxygen availability are both likely to influence P. aeruginosa colonisation and dissemination into microenvironments within the CF lung.
Conclusion
This study provides novel findings concerning the impact of anoxia upon interactions between S. aureus and CF isolates of P. aeruginosa in mixed planktonic culture and mixed species biofilms. We hypothesise from our data that anoxia is one of several potential mechanisms facilitating S. aureus-P. aeruginosa co-existence in the CF lung, with co-infection having been associated with poor clinical outcomes and a worsening of pulmonary function in CF20,90. However, not all P. aeruginosa CF isolates lose their ability to antagonise S. aureus growth under anoxia. Further work is required to determine the nature of the major anti-staphylococcal compound which facilitates P. aeruginosa dominance under both normoxia and anoxia, although this study provides evidence that the factor is likely to be heat-stable and >3 kDa in size. The ability of select P. aeruginosa CF isolates to retain their virulence and motility under anoxia is likely to influence disease progression differently across the CF population. The ability of S. aureus exoproducts to further modulate P. aeruginosa motility in an isolate dependent manner, reinforces the complex interspecies interactions known occur within the CF lung.
As co-isolated strains of S. aureus and P. aeruginosa are shown to be less antagonistic than those from mono-infected patients82, a longitudinal study of select S. aureus and P. aeruginosa co-isolates could be performed. This would allow interspecies interactions and the pathoadaptive mechanisms that occur overtime in both S. aureus and P. aeruginosa to be studied in greater detail and allow comparisons to be made under different environmental conditions. It is hoped that this study encourages interspecies interactions of other key CF pathogens to be determined under anoxia, an environmental condition shown to be important in modulating bacterial virulence and interspecies interactions.
Materials and Methods
Bacterial strains and growth conditions
All bacterial strains used in this study are listed in Table 1, with Table 2 describing their colony morphotype following growth upon LB agar. Eight clinical isolates of P. aeruginosa were cultured from paediatric CF sputum samples from Birmingham Children’s Hospital, UK between 1990–1999. All bacterial strains were stored at −80 °C in 50% (v/v) glycerol/water. Bacteria were streaked from frozen stocks to Luria Bertani (LB) agar plates (Fisher Scientific, UK) and incubated at 37 °C for 48 h. Strains were identified using selective agar, Gram staining and biochemical testing.
Table 1.
S. aureus and P. aeruginosa strains used in this study.
| Strain | Species | Source | Reference |
|---|---|---|---|
| 6538 | S. aureus | American Type Culture Collection (ATCC) | 99 |
| PAO1 | P. aeruginosa | Wound exudate, Melbourne, Australia | 100 |
| Isolate 1 | P. aeruginosa | CF Sputum from Birmingham Children’s Hospital, Birmingham, UK | This study |
| Isolate 2 | |||
| Isolate 3 | |||
| Isolate 4 | |||
| Isolate 5 | |||
| Isolate 6 | |||
| Isolate 7 | |||
| Isolate 8 |
Table 2.
Colony morphology of P. aeruginosa laboratory strain PAO1 and clinical CF isolates upon LB agar plates.
| Isolate | Pigmentation | Colony Size | Mucoidy status | Form | Optical Property | Autolysis | Surface Texture | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brown | Green | White | Opaque | Small | Large | Mucoid | Non-mucoid | Circular | Irregular | Opaque | Rough | Smooth | ||
| PAO1 | + | + | + | + | + | − | + | |||||||
| 1 | + | + | + | + | + | + | − | + | ||||||
| 2 | + | + | + | + | + | − | + | |||||||
| 3 | + | + | + | + | + | − | + | |||||||
| 4 | + | + | + | + | + | + | − | + | ||||||
| 5 | + | + | + | + | + | − | + | |||||||
| 6 | + | + | + | + | + | − | + | |||||||
| 7 | + | + | + | + | + | − | + | |||||||
| 8 | + | + | + | + | + | − | + | |||||||
The production of four colony pigmentations were studied. Colonies were positive for mucoidy status if they exhibited a slimy appearance. Colony form assessed the basic shape of the colonies. Autolysis assessed whether the bacteria grew as colonies that were lysed in their centres, whilst margin determined the edge of the colonies.
All assays were carried out using standardised bacterial inoculum. Single well isolated colonies of S. aureus or P. aeruginosa were routinely inoculated into 10 mL of sterile LBN broth (LB broth supplemented with 1% (w/v) potassium nitrate) (Fisher Scientific, UK) and grown at 37 °C under static conditions of normoxia or anoxia (miniMACS anaerobic workstation, Don Whitley Scientific, UK) for approximately 16 h.
Cross-Streak assay on solid agar
Overnight cultures of S. aureus and P. aeruginosa grown separately under normoxia or anoxia were pelleted, resuspended in fresh broth and adjusted to an OD470 of 1.0. A sterile cotton swab was immersed in a given P. aeruginosa standardised culture and streaked horizontally across the surface of a LB agar plate. After air drying for 20 min, a sterile cotton swab was immersed in the adjusted S. aureus culture and cross-streaked vertically across the surface of the agar plate. Plates were then incubated either under normoxia or anoxia at 37 °C for 18 h, prior to being visually inspected for growth inhibition.
Mono-culture and co-culture planktonic growth curves
All growth curve experiments were conducted in 250 mL conical flasks containing 50 mL of LBN broth at 37 °C under static conditions. Overnight cultures of S. aureus and P. aeruginosa CF isolates grown under normoxia or anoxia were pelleted, resuspended in fresh medium and adjusted to an OD470 of 1.0. For co-culture growth curves, the bacteria were inoculated at an equal ratio (1:1, S. aureus to P. aeruginosa) and incubated under static conditions at 37 °C for 24 h. Samples were taken at regular intervals, serially diluted in sterile PBS (Fisher, UK) and 20 µL spots plated onto Pseudomonas isolation agar (PIA) (Fisher Scientific, UK) and mannitol salt agar (MSA) (Fisher Scientific, UK) to allow species differentiation. The plates were incubated for approximately18 h, prior to enumerating the colony forming units (CFU/mL).
The competition index (CI) and Relative Increase Ratio (RIR) were calculated. The RIR was calculated on single growth curve data using the P. aeruginosa-S. aureus ratio at a given time point, divided by the same ratio at time point 0 h (inoculum). The same ratio was used to calculate the CI, although this used data from the mixed culture. A CI that differs statistically from the RIR indicates competition between the two organisms. This method was adapted from Macho et al.91.
Mono-culture and co-culture biofilm formation
Overnight cultures of S. aureus and P. aeruginosa grown under normoxia or anoxia were centrifuged and adjusted to an OD470 of 1.0. Cultures were diluted tenfold and 100 µL added to the central wells of a 96-well tissue culture treated flat bottom plate, either individually or in a 1:1 ratio for an hour under static conditions, at 37 °C. An equal volume of broth was added to the individual culture to compensate for any dilution effect. After 60 min, the well contents were aspirated and replaced with fresh LBN broth. Plates were incubated for a further 24 h at 37 °C under static conditions. Following this, biofilms were washed twice using 200 µL of PBS, detached using 100 µL of 0.25% trypsin-EDTA (Fisher Scientific, UK), collected, vortexed for 70 s, serially diluted and plated onto PIA (Fisher, UK) and MSA (Fisher Scientific, UK). The plates were incubated for approximately18 h, prior to enumerating the colony forming units (CFU/mL).
Preparation of P. aeruginosa cell-free culture supernatant
Overnight cultures of P. aeruginosa grown under normoxia or anoxia were centrifuged at 4,000 x g for 10 min at 4 °C. Each supernatant was sterile filtered with a low-binding 0.22 µm polyethersulfone membrane filter (Corning, USA) and stored at −20 °C until needed. To confirm sterility after each preparation, a small volume of supernatant was streaked onto LB agar plates and incubated for approximately 20 h prior to reading. For size exclusion experiments, 10 mL of cell-free supernatant was added to a 3 kDa molecular weight cut off protein concentrator (ThermoFisher, UK) and centrifuged at 4,000 × g for 1 h. Apical and basal volumes were subsequently added to 2.0 mL sterile microcentrifuge tubes (Fisher Scientific, UK). For heat-treatment, microcentrifuge tubes containing cell-free supernatant were added to a heat block and boiled at 95 °C for 10 mins, prior to cooling.
Determination of total protease production
Protease production was determined using skimmed milk agar. Cell-free supernatants (40 µL) from overnight cultures were loaded into wells in agar plates and incubated at 37 °C for 24 h. Hydrolysis of the milk protein casein results in a clear zone surrounding the bacterial supernatant and would show evidence of protease production. LBN medium was also loaded as a negative control. Clearance zone diameters were measured with a ruler in millimetres (mm).
Staphylolytic activity
This method was adapted from92,93. An overnight culture of S. aureus grown under static normoxia was centrifuged at 4,000 × g for 10 min at 4 °C, prior to the pellet being resuspended in 250 µL of 25 mM diethanolamine buffer (Sigma, UK), pH 9.5. The bacteria were then heated to 100 °C for 10 min, before being diluted to a final optimal density OD595 of 1.0. 400 µL of the adjusted heat-killed S. aureus was then added to each microtube (Fisher, UK). The cell-free supernatant from each P. aeruginosa isolate was diluted 1:10 with diethanolamine buffer, prior to 100 µL being added to the heat-killed S. aureus. Bacterial lysis was determined by measuring decreases in heat killed S. aureus OD595 after 60 min on a MultiSkan Go plate reader (ThermoFisher, UK). LBN broth alone was used as a control.
Pyocyanin extraction and quantification
This method was adapted from94,95. Overnight cultures of P. aeruginosa grown under normoxia or anoxia were pelleted, resuspended in fresh medium and adjusted to an OD470 of 1.0. For single cultures, 500 µL of P. aeruginosa was then added to a 250 mL conical flask containing 50 mL of LBN broth (1:100 dilution). The flasks were then incubated at 37 °C for 24 h, under static normoxia or anoxia. To quantify pyocyanin production, bacterial cells were pelleted by 4,000 × g centrifugation for 25 min at 4 °C and the supernatant sterile filtered with a low-binding 0.22 µm polyethersulfone membrane filter (Corning, USA). 7.5 mL of the sterile supernatant was added to 4.5 mL of chloroform and vortexed for ten, 2 s intervals. The sample was centrifuged at 4,000 × g for 1 min at 4 °C, prior to 3 mL of the blue-green phase (chloroform phase) being aspirated into a new tube. 1.5 mL of 0.2 M hydrochloric acid was then added to the tube and vortexed again for ten, 2 s intervals, prior to centrifugation at 4,000 × g for 1 min at 4 °C. 100 µL of the pink coloured phase was then transferred into a 96-well plate in triplicate. 100 µL of hydrochloric acid was added in triplicate as a control. The plate was then read at OD520 and multiplied by the extinction co-efficient 17.072 to determine the concentration of pyocyanin per mL of supernatant94.
Pyoverdine fluorescence spectroscopy
A total of 100 µL of each P. aeruginosa cell-free supernatant was added to a black 96-well plate (Griener Bio One, UK) in triplicate and read at excitation and emission wavelengths of 400/460 nm as performed previously96,97 on a Gemini XS Spectramax (Molecular Devices, USA) fluorescence plate reader. The background level of fluorescence was measured using 100 µL LBN broth only.
Drop collapse assay
Cell-free supernatants from overnight cultures of P. aeruginosa were serially diluted (1:1) in sterile dH2O containing 0.005% crystal violet to aid visualisation. A total of 20 µL of each dilution (including neat supernatant) were spotted onto the underside of a lid of a 96-well plate and the plate titled at 90°, as performed previously82. The assay works on the principle that if the droplet contains surfactants, the drops spread. However, as the quantity of surfactants decrease by dilution, the droplet eventually beads up due to an increase in surface tension. Surfactant scores are equal to the reciprocal of the greatest dilution at which there was surfactant activity (a collapsed drop).
Secretome analysis
Pooled overnight cultures of P. aeruginosa grown under either normoxia or anoxia were pelleted following centrifugation at 4,000 × g for 30 min at 4 °C. The supernatant containing extracellular proteins was sterile filtered through a 0.22 µm polyethersulfone membrane filter (Corning, USA). Supernatants were concentrated using Amicon 3 kDa cut-off filters (Millipore, UK) prior to precipitation with 25% trichloroacetic acid (TCA; w/v) for 15 min on ice. Proteins were pelleted at 14000 × g for 10 min, and pellets washed with acetone. Protein pellets were solubilised in 50 mM pH 7.4 Tris-HCl containing 2 mM CHAPS, 7 M urea and 7 M thiourea using an ultrasonicating probe (30 s sonication per cycle, 65% full power, 2 cycles), and quantified against a BSA calibration curve using the Bradford assay (BioRad, UK). Proteins (30 µg) were reduced in Laemmli buffer (Sigma Aldrich, UK) for 15 min at 65 °C, and separated by SDS-PAGE on a 10% gel. Separated proteins were stained with Coomassie G250 blue (0.5% w/v in 40% aqueous methanol and 10% glacial acetic acid) for 1 h and destained in an aqueous solution of 10% ethanol and 7.5% glacial acetic acid.
Protein in-gel digestion was performed following the method of Schevchenko et al.98, with minor modifications. Each sample lane was divided into 5 bands, gel bands were excised and diced in a clean polypropylene tube using a sterile scalpel. Gel pieces were subsequently destained with 50% acetonitrile in 50 mM ammonium bicarbonate, dehydrated with acetonitrile and vacuum dried. Proteins were in-gel digested using trypsin in 3 mM ammonium bicarbonate (25:1 protein to trypsin ratio; Trypsin Gold, sequencing, Promega, UK) coupled with shaking at 550 rpm at 37 °C overnight. Peptides were extracted for 15 min in an ultrasonic bath initially using a volume of acetonitrile equivalent to 50% of sample volume, two times with 150 µL of 50% acetonitrile in 50 mM ammonium bicarbonate. Finally 400 µL of pure acetonitrile was used to fully dehydrated the gel pieces and maximise peptide extraction. Each time the complete peptide extract was collected in a polypropylene tube. Finally, peptide extracts were vacuum dried and stored at −20 °C prior to analysis.
Samples were reconstituted in 50 µL of 3% aqueous acetonitrile and 0.1% formic acid for liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) analysis. Peptides were separated and analysed using an nLC system (Dionex 3000, ThermoScientific, UK) coupled to 5600 TripleTof (AB Sciex, UK) operating in information dependent (IDA) mode. Peptide solution (10 µL, ~5 pmol) was injected onto a trap column (PepMapTM, C18, 5 µm, 100 Å, 300 µm × 1 mm, ThermoScientific, UK) using 2% of eluent B (98% acetonitrile in aqueous 0.1% formic acid) at a flow rate of 30 µL/min. Peptides were subsequently separated on an analytical column (AcclaimTM, PepMapTM C18, 3 µm, 100 Å, 75 µm × 150 mm, ThermoScientific, UK) with the following gradient: 0–3 min 2% B, 3–48 min 2–45% B, 48–52 min 45–90% B, 52–55 min 90% B, 55–70 min 2% B). Electrospray was formed by spraying the nLC eluate at 2500 V using a PicoTipTM emitter (New Objective, Germany). The 10 most intense ions from each MS survey scan were selected for MS/MS, while acquired ions were temporarily excluded from MS/MS acquisition for 30 s. The mass spectrometer was calibrated prior to acquisition to ensure a high mass accuracy (<10 ppm) on both MS and tandem mass spectrometry (MS/MS) levels. Relative quantification was done using Progenesis QI for proteomics software (version 4, Nonlinear Dynamics, UK). Only protein unique peptides were used for relative quantification. MS/MS data were searched using MasconDeamon (ver 2.5) against the SwissProt database, with the following search restriction parameters: mass tolerance of 0.1 Da for MS and 0.6 Da for MS/MS spectra, a maximum of 2 trypsin miscleavages, Pseudomonas aeruginosa taxonomy, variable modifications of methionine oxidation and cysteine carbamidomethylation.
Bacterial motility
Swimming motility of P. aeruginosa was investigated using 0.3% (w/v) nutrient agar plates supplemented with nutrient broth and 1% (w/v) potassium nitrate. Swarming of P. aeruginosa was determined using 0.5% (w/v) nutrient agar plates supplemented with nutrient broth, dextrose and 1% (w/v) potassium nitrate. Overnight cultures of P. aeruginosa grown under normoxia or anoxia were adjusted to an OD470 of 1.0 with a total of 5 µL of culture added to the centre of each plate. To measure the effects of S. aureus on P. aeruginosa motility, a 1:100 diluted S. aureus cell-free culture supernatant was added to the agar plate. Plates were incubated under normoxia or anoxia for 24 h at 37 °C. The diameter of the zone travelled by P. aeruginosa was then measured using a ruler in mm.
Statistical analysis
All results unless otherwise stated are expressed as mean ± S.E.M. Data for each experiment were collected from three independent experiments (N = 3), each performed in triplicate. All statistical analyses were performed using GraphPad Prism 6 software (Graphpad, La Jolla, CA, USA) with significance being set to P < 0.05. The specific tests used for each experiment are described in the figure legends.
Supplementary information
Acknowledgements
We gratefully acknowledge the Humane Research Trust for funding LJM and RP for this research. We also acknowledge support for IM and AD from BBSRC (BB/M006298/1). We also gratefully acknowledge the contributions of Roberta Liccardo, Nikita Karra and Tafadzwa Manaka for their technical support.
Author Contributions
R.P., P.A.L., A.D. and L.J.M. designed the study. P.A.L. acquired the C.F. clinical isolates. R.P. performed the experiments, whilst R.P. and A.D. analysed and interpreted the data. I.M. performed the mass spectrometry and analysed the data obtained. R.P. prepared the figures and drafted the manuscript. All authors edited the manuscript.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42952-x.
References
- 1.Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Welsh MJ, Smith AE. Molecular Mechanisms of Cftr Chloride Channel Dysfunction in Cystic-Fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-R. [DOI] [PubMed] [Google Scholar]
- 3.Matsui H, et al. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell. 1998;95:1005–1015. doi: 10.1016/S0092-8674(00)81724-9. [DOI] [PubMed] [Google Scholar]
- 4.Kreda SM, Davis CW, Rose MC. CFTR, mucins, and mucus obstruction in cystic fibrosis. Cold Spring Harbor perspectives in medicine. 2012;2:a009589. doi: 10.1101/cshperspect.a009589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams RE. Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev. 1963;27:56–71. doi: 10.1128/br.27.1.56-71.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahl BC. Impact of Staphylococcus aureus on the pathogenesis of chronic cystic fibrosis lung disease. Int J Med Microbiol. 2010;300:514–519. doi: 10.1016/j.ijmm.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Trust, C. F. UK Cystic Fibrosis Registry Annual Data Report 2016 (2016).
- 8.Burns JL, et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 9.Kidd TJ, et al. Shared Pseudomonas aeruginosa genotypes are common in Australian cystic fibrosis centres. Eur Respir J. 2013;41:1091–1100. doi: 10.1183/09031936.00060512. [DOI] [PubMed] [Google Scholar]
- 10.Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J. 2012;40:227–238. doi: 10.1183/09031936.00204411. [DOI] [PubMed] [Google Scholar]
- 11.Cox MJ, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rau MH, et al. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ Microbiol. 2010;12:1643–1658. doi: 10.1111/j.1462-2920.2010.02211.x. [DOI] [PubMed] [Google Scholar]
- 13.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folkesson A, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 15.Hogardt M, Heesemann J. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol. 2010;300:557–562. doi: 10.1016/j.ijmm.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Kessler E, Safrin M, Olson JC, Ohman DE. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem. 1993;268:7503–7508. [PubMed] [Google Scholar]
- 17.Voggu L, et al. Microevolution of cytochrome bd oxidase in Staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol. 2006;188:8079–8086. doi: 10.1128/JB.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biswas L, Biswas R, Schlag M, Bertram R, Gotz F. Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl Environ Microbiol. 2009;75:6910–6912. doi: 10.1128/AEM.01211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother. 1992;30:615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 20.Limoli DH, et al. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur J Clin Microbiol Infect Dis. 2016;35:947–953. doi: 10.1007/s10096-016-2621-0. [DOI] [PubMed] [Google Scholar]
- 21.Wakeman CA, et al. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat Commun. 2016;7:11951. doi: 10.1038/ncomms11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogan DA, et al. Analysis of Lung Microbiota in Bronchoalveolar Lavage, Protected Brush and Sputum Samples from Subjects with Mild-To-Moderate Cystic Fibrosis Lung Disease. PLoS One. 2016;11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Com G, et al. Predictors and outcome of low initial forced expiratory volume in 1 second measurement in children with cystic fibrosis. J Pediatr. 2014;164:832–838. doi: 10.1016/j.jpeds.2013.11.064. [DOI] [PubMed] [Google Scholar]
- 24.Kessler E, Safrin M, Olson JC, Ohman DE. Secreted Lasa of Pseudomonas-Aeruginosa Is a Staphylolytic Protease. Journal of Biological Chemistry. 1993;268:7503–7508. [PubMed] [Google Scholar]
- 25.Baldan, R. et al. Adaptation of Pseudomonas aeruginosa in Cystic Fibrosis Airways Influences Virulence of Staphylococcus aureus In Vitro and Murine Models of Co-Infection. Plos One9, 10.1371/journal.pone.0089614 (2014). [DOI] [PMC free article] [PubMed]
- 26.Fugere, A. et al. Interspecific Small Molecule Interactions between Clinical Isolates of Pseudomonas aeruginosa and Staphylococcus aureus from Adult Cystic Fibrosis Patients. Plos One9, 10.1371/journal.pone.0086705 (2014). [DOI] [PMC free article] [PubMed]
- 27.Filkins LM, et al. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards Fermentative Metabolism and Reduced Viability in a Cystic Fibrosis Model. J Bacteriol. 2015;197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fothergill, J. L. et al. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. Bmc Microbiol7, 10.1186/1471-2180-7-45 (2007). [DOI] [PMC free article] [PubMed]
- 29.Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187:554–566. doi: 10.1128/Jb.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanderson K, Wescombe L, Kirov SM, Champion A, Reid DW. Bacterial cyanogenesis occurs in the cystic fibrosis lung. Eur Respir J. 2008;32:329–333. doi: 10.1183/09031936.00152407. [DOI] [PubMed] [Google Scholar]
- 31.Worlitzsch D, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon SS, et al. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell. 2002;3:593–603. doi: 10.1016/S1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 33.Schobert M, Jahn D. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol. 2010;300:549–556. doi: 10.1016/j.ijmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Stutts MJ, Knowles MR, Gatzy JT, Boucher RC. Oxygen consumption and ouabain binding sites in cystic fibrosis nasal epithelium. Pediatr Res. 1986;20:1316–1320. doi: 10.1203/00006450-198612000-00026. [DOI] [PubMed] [Google Scholar]
- 35.Sherrard LJ, Bell SC, Tunney MM. The role of anaerobic bacteria in the cystic fibrosis airway. Curr Opin Pulm Med. 2016;22:637–643. doi: 10.1097/MCP.0000000000000299. [DOI] [PubMed] [Google Scholar]
- 36.Tunney MM, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med. 2008;177:995–1001. doi: 10.1164/rccm.200708-1151OC. [DOI] [PubMed] [Google Scholar]
- 37.Harris JK, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costerton JW. Anaerobic biofilm infections in cystic fibrosis. Mol Cell. 2002;10:699–700. doi: 10.1016/S1097-2765(02)00698-6. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber K, et al. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol. 2007;189:4310–4314. doi: 10.1128/JB.00240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassett DJ. Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts diffusion of oxygen. J Bacteriol. 1996;178:7322–7325. doi: 10.1128/jb.178.24.7322-7325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son MS, Matthews WJ, Jr., Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beckmann C, et al. Use of phage display to identify potential Pseudomonas aeruginosa gene products relevant to early cystic fibrosis airway infections. Infect Immun. 2005;73:444–452. doi: 10.1128/IAI.73.1.444-452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang H, et al. The impact of anaerobiosis on strain-dependent phenotypic variations in Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 2013;77:1747–1752. doi: 10.1271/bbb.130309. [DOI] [PubMed] [Google Scholar]
- 44.Lee KM, Yoon MY, Park Y, Lee JH, Yoon SS. Anaerobiosis-induced loss of cytotoxicity is due to inactivation of quorum sensing in Pseudomonas aeruginosa. Infect Immun. 2011;79:2792–2800. doi: 10.1128/IAI.01361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaines JM, Carty NL, Colmer-Hamood JA, Hamood AN. Effect of static growth and different levels of environmental oxygen on toxA and ptxR expression in the Pseudomonas aeruginosa strain PAO1. Microbiology. 2005;151:2263–2275. doi: 10.1099/mic.0.27754-0. [DOI] [PubMed] [Google Scholar]
- 46.Zimmermann A, Reimmann C, Galimand M, Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991;5:1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 47.Bragonzi A, et al. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J Infect Dis. 2005;192:410–419. doi: 10.1086/431516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borriello G, et al. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother. 2004;48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tote K, Horemans T, Vanden Berghe D, Maes L, Cos P. Inhibitory effect of biocides on the viable masses and matrices of Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2010;76:3135–3142. doi: 10.1128/AEM.02095-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park JH, Lee JH, Cho MH, Herzberg M, Lee J. Acceleration of protease effect on Staphylococcus aureus biofilm dispersal. FEMS Microbiol Lett. 2012;335:31–38. doi: 10.1111/j.1574-6968.2012.02635.x. [DOI] [PubMed] [Google Scholar]
- 51.Makarova, O., Johnston, P., Walther, B., Rolff, J. & Roesler, U. Complete Genome Sequence of the Disinfectant Susceptibility Testing Reference Strain Staphylococcus aureus subsp. aureus ATCC 6538. Genome Announc5, 10.1128/genomeA.00293-17 (2017). [DOI] [PMC free article] [PubMed]
- 52.LaFayette, S. L. et al. Cystic fibrosis-adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Sci Adv1, 10.1126/sciadv.1500199 (2015). [DOI] [PMC free article] [PubMed]
- 53.Caballero AR, et al. Pseudomonas aeruginosa protease IV enzyme assays and comparison to other Pseudomonas proteases. Anal Biochem. 2001;290:330–337. doi: 10.1006/abio.2001.4999. [DOI] [PubMed] [Google Scholar]
- 54.Oldak E, Trafny EA. Secretion of proteases by Pseudomonas aeruginosa biofilms exposed to ciprofloxacin. Antimicrob Agents Chemother. 2005;49:3281–3288. doi: 10.1128/AAC.49.8.3281-3288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Brien S, et al. High virulence sub-populations in Pseudomonas aeruginosa long-term cystic fibrosis airway infections. BMC Microbiol. 2017;17:30. doi: 10.1186/s12866-017-0941-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hassett DJ, et al. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 2009;17:130–138. doi: 10.1016/j.tim.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 57.Palmer KL, Mashburn LM, Singh PK, Whiteley M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yadav V, Harjai K, Kaur R, Joshi K, Sharma S. Urovirulence of Pseudomonas aeruginosa: planktonic cells vs. biofilm cells. Folia Microbiol (Praha) 2004;49:465–470. doi: 10.1007/BF02931610. [DOI] [PubMed] [Google Scholar]
- 59.Secor PR, et al. Staphylococcus aureus Biofilm and Planktonic cultures differentially impact gene expression, mapk phosphorylation, and cytokine production in human keratinocytes. BMC Microbiol. 2011;11:143. doi: 10.1186/1471-2180-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waite RD, Papakonstantinopoulou A, Littler E, Curtis MA. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J Bacteriol. 2005;187:6571–6576. doi: 10.1128/JB.187.18.6571-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schertzer JW, Brown SA, Whiteley M. Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol Microbiol. 2010;77:1527–1538. doi: 10.1111/j.1365-2958.2010.07303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaible B, et al. Hypoxia Reduces the Pathogenicity of Pseudomonas aeruginosa by Decreasing the Expression of Multiple Virulence Factors. J Infect Dis. 2017;215:1459–1467. doi: 10.1093/infdis/jix139. [DOI] [PubMed] [Google Scholar]
- 63.Barequet IS, Ben Simon GJ, Safrin M, Ohman DE, Kessler E. Pseudomonas aeruginosa LasA protease in treatment of experimental staphylococcal keratitis. Antimicrob Agents Ch. 2004;48:1681–1687. doi: 10.1128/Aac.48.5.1681-1687.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Filiatrault MJ, et al. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect Immun. 2005;73:3764–3772. doi: 10.1128/IAI.73.6.3764-3772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conti SF, Jacobs NJ, Gray CT. Ultrastructure and respiratory capacity of Staphylococcus and Bacillus grown under aerobic and anaerobic conditions. J Bacteriol. 1968;96:554–556. doi: 10.1128/jb.96.2.554-556.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falcon MA, Mansito TB, Carnicero A, Gutierrez-Navarro AM. L-form-like colonies of Staphylococcus aureus induced by an extracellular lytic enzyme from Pseudomonas aeruginosa. J Clin Microbiol. 1989;27:1650–1654. doi: 10.1128/jcm.27.7.1650-1654.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Makino T. Induction of L-phase variant from protoplast of Staphylococcus aureus. Microbiol Immunol. 1983;27:749–755. doi: 10.1111/j.1348-0421.1983.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 68.Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013;21:73–81. doi: 10.1016/j.tim.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 70.Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. Iron Depletion Enhances Production of Antimicrobials by Pseudomonas aeruginosa. J Bacteriol. 2015;197:2265–2275. doi: 10.1128/JB.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haba E, et al. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol Bioeng. 2003;81:316–322. doi: 10.1002/bit.10474. [DOI] [PubMed] [Google Scholar]
- 72.Bharali P, Saikia JP, Ray A, Konwar BK, Rhamnolipid RL. from Pseudomonas aeruginosa OBP1: a novel chemotaxis and antibacterial agent. Colloids Surf B Biointerfaces. 2013;103:502–509. doi: 10.1016/j.colsurfb.2012.10.064. [DOI] [PubMed] [Google Scholar]
- 73.Sotirova AV, Spasova DI, Galabova DN, Karpenko E, Shulga A. Rhamnolipid-biosurfactant permeabilizing effects on gram-positive and gram-negative bacterial strains. Curr Microbiol. 2008;56:639–644. doi: 10.1007/s00284-008-9139-3. [DOI] [PubMed] [Google Scholar]
- 74.Castric PA. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can J Microbiol. 1975;21:613–618. doi: 10.1139/m75-088. [DOI] [PubMed] [Google Scholar]
- 75.Blumer C, Haas D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol. 2000;173:170–177. doi: 10.1007/s002039900127. [DOI] [PubMed] [Google Scholar]
- 76.Lightbown JW, Jackson FL. Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem J. 1956;63:130–137. doi: 10.1042/bj0630130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lepine F, Milot S, Deziel E, He J, Rahme LG. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom. 2004;15:862–869. doi: 10.1016/j.jasms.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 78.Toyofuku M, et al. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J Bacteriol. 2008;190:7947–7956. doi: 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michelsen CF, et al. Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J Bacteriol. 2014;196:3903–3911. doi: 10.1128/JB.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhagirath AY, et al. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med. 2016;16:174. doi: 10.1186/s12890-016-0339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bragonzi A, et al. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS One. 2012;7:e52330. doi: 10.1371/journal.pone.0052330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Limoli, D. H. et al. Pseudomonas aeruginosa Alginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection. MBio8, 10.1128/mBio.00186-17 (2017). [DOI] [PMC free article] [PubMed]
- 83.Beaudoin T, et al. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes. 2017;3:25. doi: 10.1038/s41522-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Orazi, G. & O’Toole, G. A. Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection. MBio8, 10.1128/mBio.00873-17 (2017). [DOI] [PMC free article] [PubMed]
- 85.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuchma SL, et al. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 88.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baraquet C, Harwood CS. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci USA. 2013;110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maliniak ML, Stecenko AA, McCarty NA. A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: A single-center study. J Cyst Fibros. 2016;15:350–356. doi: 10.1016/j.jcf.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 91.MACHO ALBERTO P., ZUMAQUERO ADELA, ORTIZ-MARTÍN INMACULADA, BEUZÓN CARMEN R. Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae?plant interactions. Molecular Plant Pathology. 2007;8:437–450. doi: 10.1111/j.1364-3703.2007.00404.x. [DOI] [PubMed] [Google Scholar]
- 92.Kong KF, et al. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob Agents Chemother. 2005;49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grande KK, Gustin JK, Kessler E, Ohman DE. Identification of critical residues in the propeptide of LasA protease of Pseudomonas aeruginosa involved in the formation of a stable mature protease. J Bacteriol. 2007;189:3960–3968. doi: 10.1128/JB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu D, Huang W, Duan Q, Li F, Cheng H. Sodium houttuyfonate affects production of N-acyl homoserine lactone and quorum sensing-regulated genes expression in Pseudomonas aeruginosa. Front Microbiol. 2014;5:635. doi: 10.3389/fmicb.2014.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Andersen SB, Marvig RL, Molin S, Krogh Johansen H, Griffin AS. Long-term social dynamics drive loss of function in pathogenic bacteria. Proc Natl Acad Sci USA. 2015;112:10756–10761. doi: 10.1073/pnas.1508324112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krzyzanowska DM, et al. When Genome-Based Approach Meets the “Old but Good”: Revealing Genes Involved in the Antibacterial Activity of Pseudomonas sp. P482 against Soft Rot Pathogens. Front Microbiol. 2016;7:782. doi: 10.3389/fmicb.2016.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 99.Forbes S, Latimer J, Bazaid A, McBain AJ. Altered Competitive Fitness, Antimicrobial Susceptibility, and Cellular Morphology in a Triclosan-Induced Small-Colony Variant of Staphylococcus aureus. Antimicrob Agents Chemother. 2015;59:4809–4816. doi: 10.1128/AAC.00352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Holloway BW. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.