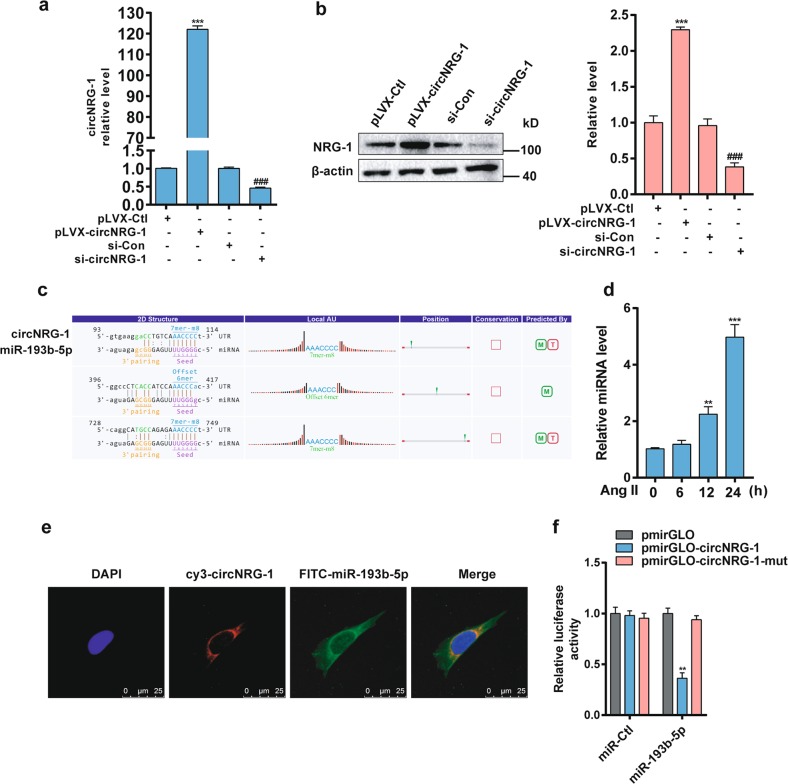
Fig. 3. circNRG-1 acts as a miR-193b-5p sponge to regulate NRG-1 expression.
a MASMCs were transfected with pLVX-circNRG-1 or si-circNRG-1, as well as with their corresponding controls. circNRG-1 expression was analyzed by qRT-PCR. ***P < 0.001 vs. pLVX-Ctl, ###P < 0.001 vs. si-Con. b MASMCs were transfected with pLVX-Ctl, pLVX-circNRG-1, si-Con and si-circNRG-1 for 24 h. NRG-1 was quantitated by densitometry and values were normalized to total β-actin (Right panel). Data were means ± SEM, n = 3. ***P < 0.001 vs. pLVX-Ctl, ###P < 0.001 vs. si-Con. c miR-193b-5p binding sites in the circNRG-1 sequence were predicted. d miR-193b-5p level in MASMCs treated with Ang II (10−7 M) for the indicated times was examined by qRT-PCR. Data are presented as means ± SEM, n = 3. **P < 0.01, ***P < 0.001 vs. 0 h. e Co-localization between circNRG-1 and miR-193b-5p was detected by RNA in situ hybridization in MASMCs. Nuclei were stained with DAPI. Scale bars = 25 μm. f HEK 293A cells were co-transfected with wild-type pmirGLO-circNRG-1 or its mutant pmirGLO-circNRG-1 mut and miR-193b-5p mimic or miR-Ctl. Luciferase activity was measured. Data are the means ± SEM of three independent experiments. **P < 0.01 vs. pmirGLO, pmirGLO-circNRG-1 mut