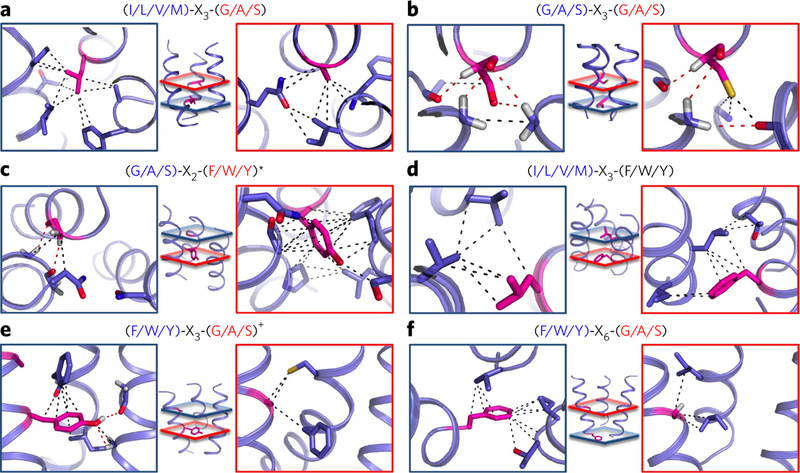
Figure 3. Consensus patterns of contacts display unique combinations of atomic interactions.
(a−f) Enriched sequence motifs create unique trimer-specific interatomic van der Waals (black dotted line) and hydrogen bonding (red dotted line) interactions in local regions of the trimers defined by red and blue planes at the center of each panel. Interatomic interactions are defined for any pair of atoms distant by <5 Å belonging to one of the two motif residues (magenta) and a residue on an adjacent helix. The red and blue boxes highlight the atomic contacts from the corresponding colored planes created by each residue of the following motifs and trimer structure classes: (G/A/S)-X3-(G/A/S) motif specific to all right-handed trimers (a); (I/L/V/M)-X3-(G/A/S) motif specific to all left-handed trimers (b); (G/A/S)-X2-(F/W/Y/(M)) motif specific to parallel and left-handed trimers (c); (I/L/V/M)-X3-(F/W/Y) motif specific to parallel and right-handed trimers (d); (F/W/Y)-X3-(G/A/S) motif specific to left- and right-handed trimers (e); (F/W/Y)-X6-(G/A/S) motif specific to left- and right-handed IItrimers (f). *, methionine also found in the second position with similar contacts; +, histidine also found in the first position with similar contacts.