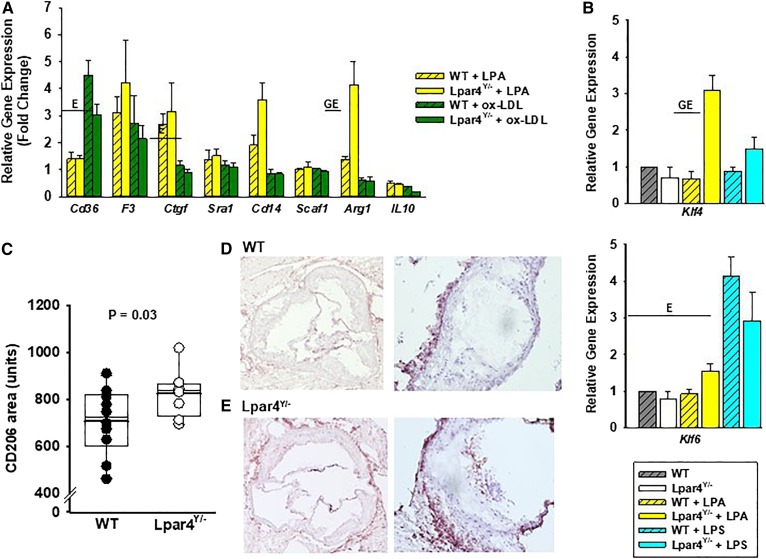
Fig. 3.
LPAR4 deficiency influences oxidized LDL-elicited gene expression in BMDMs. A: Gene expression analysis in cells isolated from WT (open bars) or Lpar4Y/− (hatched bars) mice. Cells were harvested from four mice, cultured for 7 days, and then exposed to 18:1 LPA (5 μM; yellow bars), ox-LDL (50 μg/ml; green bars), or vehicle control for 3 h. Results are presented relative to the expression with vehicle exposure and are summarized from three separate experiments analyzed by two-way ANOVA with the Mann-Whitney test. E = significant differences in gene expression between environmental exposures (P < 0.05). GE = significant differences in gene expression based on genotype and environmental exposure (P < 0.05). B: Klf4 and Klf6 gene expression analysis in BMDMs isolated from WT (gray hatched bars; n = 4) or Lpar4Y/− (open bars; n = 4) exposed to vehicle control, 18:1 LPA (2 μM; yellow bars), or LPS (100 ng/ml; blue bars) for 3 h. Results are presented relative to expression with vehicle exposure and are summarized from three separate experiments analyzed by two-way ANOVA with the Mann-Whitney test. GE = significant differences in gene expression based on genotype and environmental exposure (P < 0.05). C: CD206+ staining in aortic root sections from WT (dark circles; n = 10) or Lpar4Y/− (open circles; n = 8) mice. Individual values represent the average of six measurements taken 80 μM apart (P = 0.03). D: Representative images of sections of WT aortic root stained for CD205 at 4× (left) and 20× (right). E: Representative images of sections of Lpar4Y/− aortic root stained for CD205 at 4× (left) and 20× (right). LPS, lipopolysaccharide.