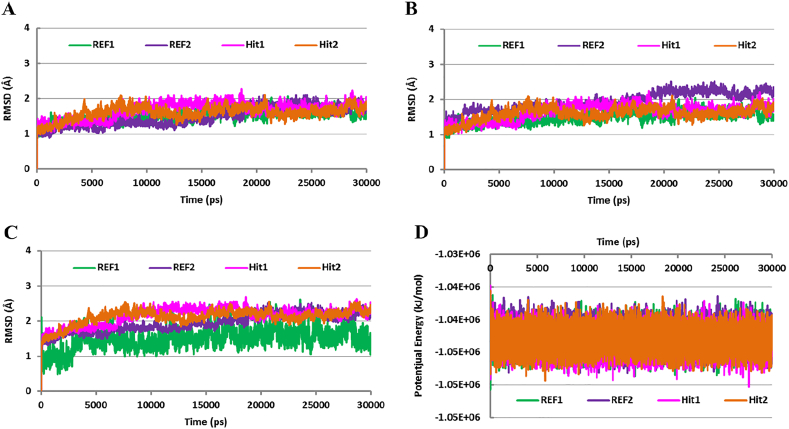
Fig. 5.
Root mean square deviation (rmsd) and potential energy analysis. The rmsd profile of the Cα atoms of Cdk5/p25 (A) and the backbone atoms of Cdk5/p25 (B). All the systems (REF1, REF2, Hit1, and Hit2) converged after 5 ns simulation and remained stable throughout the simulation period. C) Illustrates the rmsd profile of each candidate inhibitor (REF1, REF2, Hit1, and Hit2) in complex with Cdk5/p25. REF1, REF2, Hit1, and Hit2 showed stable rmsd (< 3.0 Å) after 5 ns simulation in the ATP-binding site of Cdk5/p25. D) Estimates the potential energy of each simulation system. Each system is represented by the presence of the corresponding inhibitor. Green, blue, magenta, and orange colors reflect the presence of REF1, REF2, Hit1, and Hit2, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)