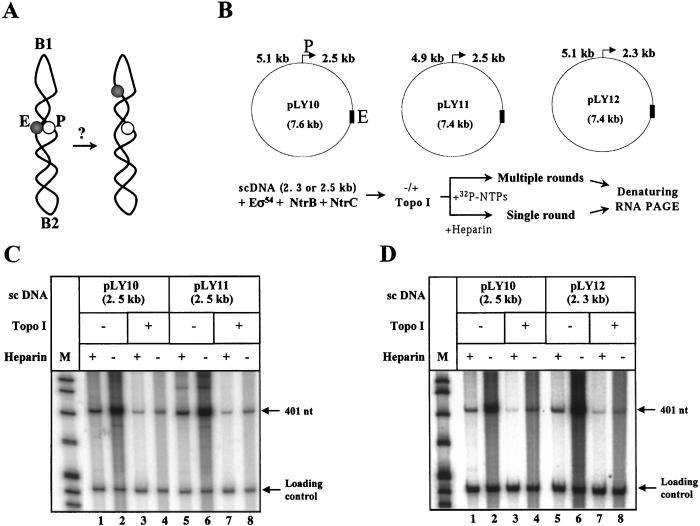
Figure 4.
Efficient activation of transcription over 2.5 kb is not a result of serendipitous enhancer–promoter juxtaposition. (A) Schematic of the experiment. Enhancer (E) and promoter (P) could be serendipitously positioned next to each other in sc DNA topologically organized by sequence-specific DNA bend(s) (B1 or B2; ref. 30). In this case, one of two asymmetrical deletions of ≈200-bp DNA on either side of the enhancer would be expected to spatially separate enhancer from promoter and decrease activity of the glnAp2 promoter on the sc template. (B) Setup of the experiment. Asymmetric ≈200-bp deletions were introduced into pLY10 on either side of the enhancer to obtain pLY11 and pLY12 plasmids. Design of the experiment is described in Fig. 2A legend and shown at the bottom. (C) Analysis of efficiency of transcription of sc and relaxed pLY11 template. (D) Analysis of efficiency of transcription of sc and relaxed pLY12 template. C and D are labeled as in Fig. 2A. pLY10 was used as a control template in the same experiment.