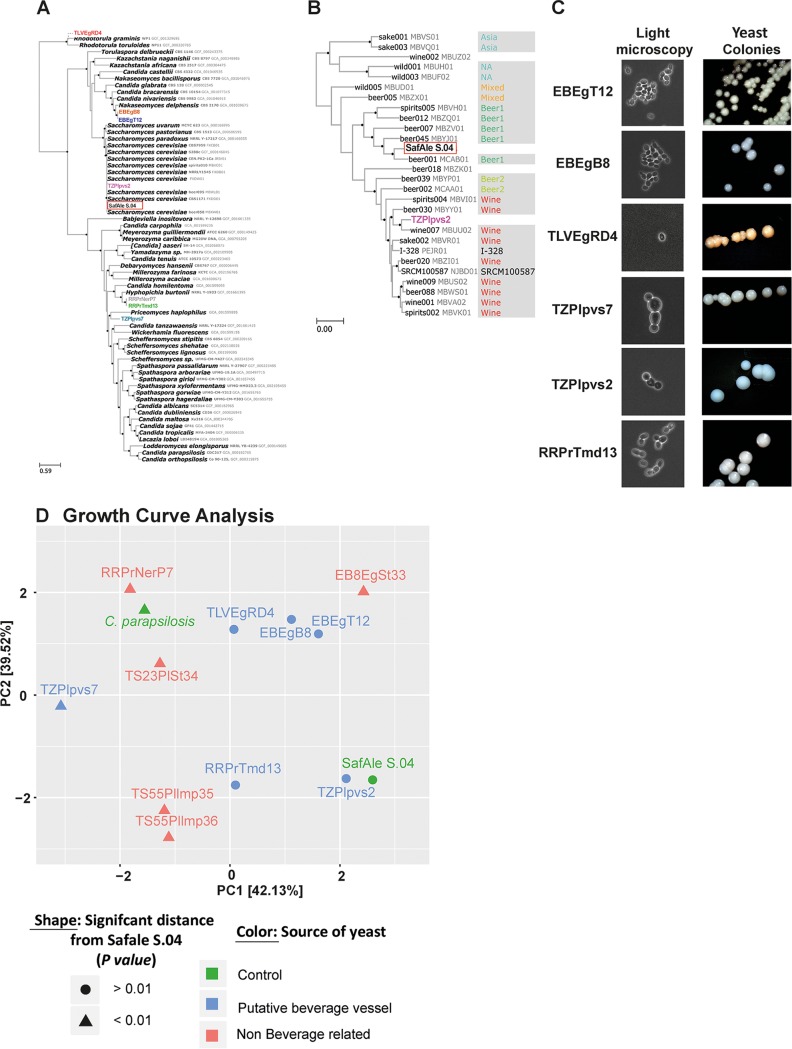
FIG 3.
Genotypic and phenotypic characterization of yeast strains isolated from ancient vessels. (A and B) Phylogenetic trees based on full-genome sequencing of the isolated yeast strains. Black bullets at nodes represent maximal bootstrap percentage of node support. The newly isolated strains are in color. The modern beer yeast species Saccharomyces cerevisiae (SafAle S.04), which served as a control, is surrounded by a red box. (A) A list of 118 gene partitions and a representation of the combination Saccharomycetaceae plus Debaryomycetaceae. (B) Comparison of TZPlpvs2 (in purple) to modern wine and beer strains, based on 465 gene representatives and partitioning of Saccharomyces cerevisiae. Reference strains are denoted by NCBI strain name and accession number followed by clade affiliation (41). (C) The shape of the isolated yeast cells under light microscopy (left panel) and colonies on YPD agar plates (right panel). (D) Growth curve analysis. The yeast strain isolated from putative beverage vessels grows in beer wort with similar kinetics to a modern beer yeast. Principal-component analysis (PCA) of the distances of the growth curves of yeast grown in beer wort under fermentation-related conditions was performed (Fig. S3). The modern domesticated beer yeast strain SafAle S.04 served as a positive control, and the pathogenic yeast C. parapsilosis served as a negative control. The marker’s shape denotes the statistical significance of the distance from SafAle S.04 growth curve kinetics, and the color denotes the source of the yeast: control, putative beverage container, or non-beverage-related vessel.